Dr. Celia Hacker, Dr. Stefania Ebli, Prof. Kathryn Hess
This repository contains the code necessary to run simulations of a specific type of Morse theory approach, called parametric Morse theory. This is done in the context of a semester project under the supervision of Celia Hacker and Stefania Ebli, in the Laboratory for Topology and Neuroscience at EPFL, directed by Kathryn Hess.
Discrete Morse theory (DMT) is the discrete analog of smooth Morse theory, as developed by Robin Forman (cf. [1]). It provides tools for analyzing the topological changes of a simplicial or cubical complex through a filtration. In brief, one can build a discrete Morse function on a given complex (a real-valued labeling of all simplices or cubes that satisfies some hierarchy conditions) and study the related so-called critical cells in the complex. Those are crucial cells in the sense that, as opposed to other cells, deleting them from the complex will fundamentally change the overall topology of the space. Generally speaking, DMT has broad applications, notably in homology computation and, more recently, in image processing (parametric Morse theory (cf. [3])).
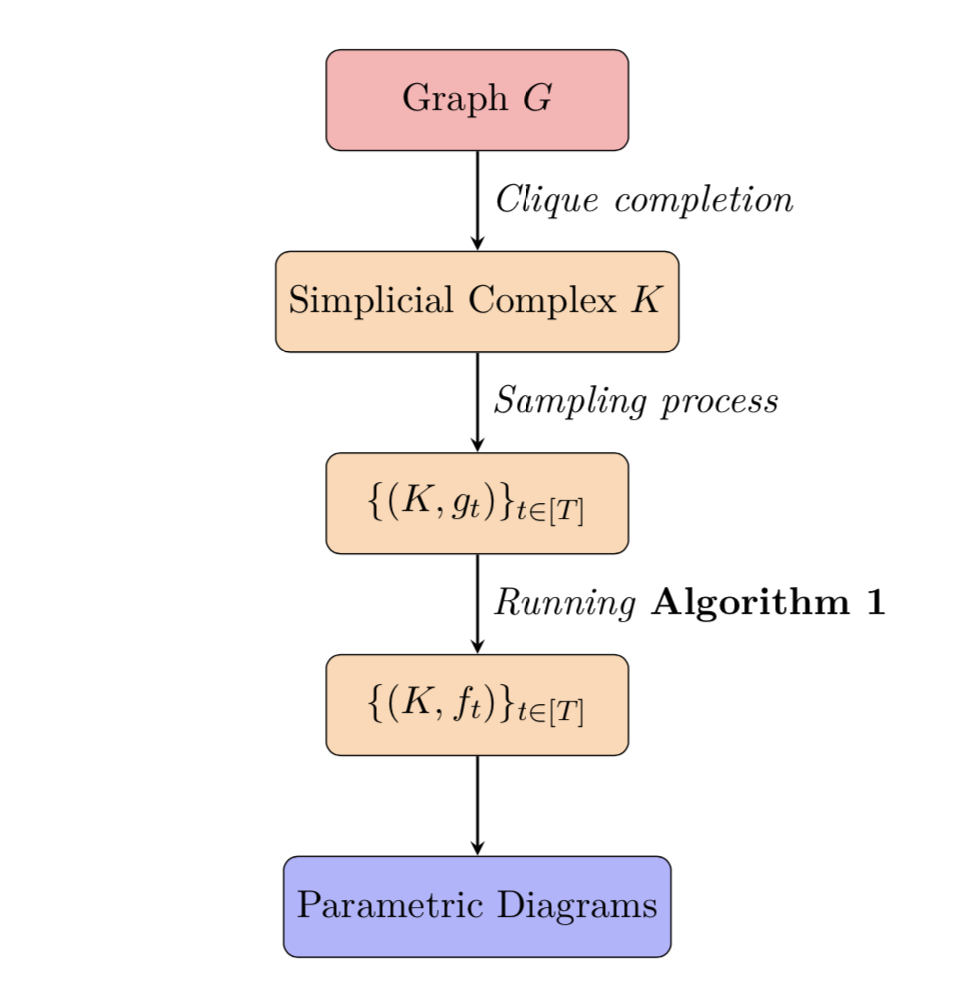
More precisely, we follow a simple pipeline described here. We start from a graph G with a mapping that assigns a continuous real-valued function with real domain to each node in G. First, we apply clique completion to the graph G to obtain a simplicial complex K. Then, we go through a sampling process to get a real value on each vertex. This "real-valued node labeling" is extended to a discrete Morse function on the whole complex K (cf. [2]). The extension algorithm is designed in the paper Persistent homology of unweighted complex networks via discrete Morse theory. Finally, we plot the parametric diagrams of the resulting finite Morse parametrization. Those diagrams encode a lot of information on the time-related behavior of critical cells that appear along the sequence of discrete Morse functions.
The src folder contains files where all the pipeline is implemented.
The overall simplified pipeline is illustrated below.
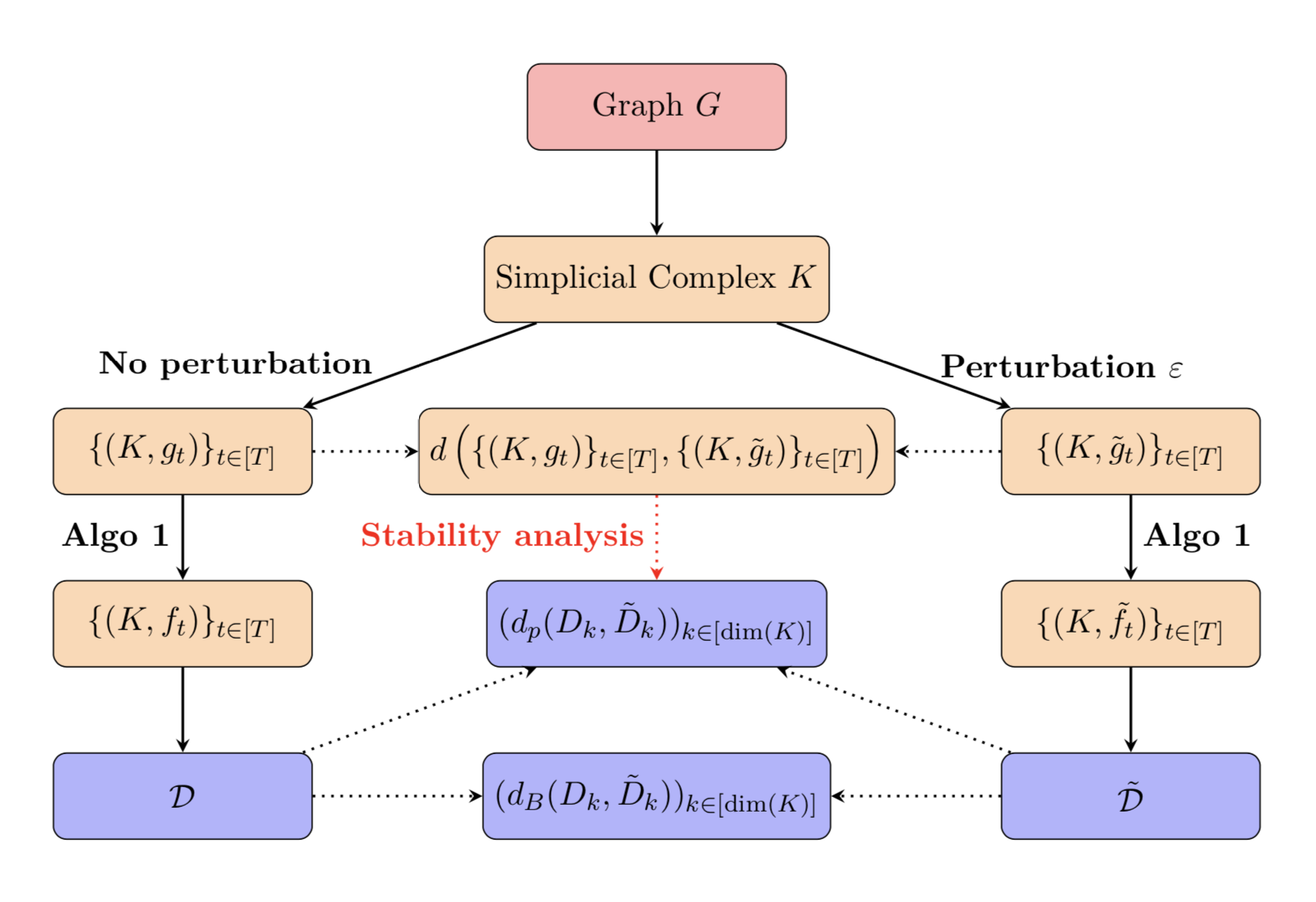
We further investigate the overall stability of the process outputting parametric diagrams. More precisely, we introduce specific distances and study the impact a perturbation on the initial sampling process has on the final output. We illustrate this in the figure below.
Use the following command to create a virtual environment and install python dependencies:
make
You can activate the environment by running:
source venv/bin/activate
and stop the environment by running:
deactivate