The Breathing K-Means is an approximation algorithm for the k-means problem that (on average) is better (higher solution quality) and faster (lower CPU time usage) than k-means++.
Techreport: https://arxiv.org/abs/2006.15666 (submitted for publication)
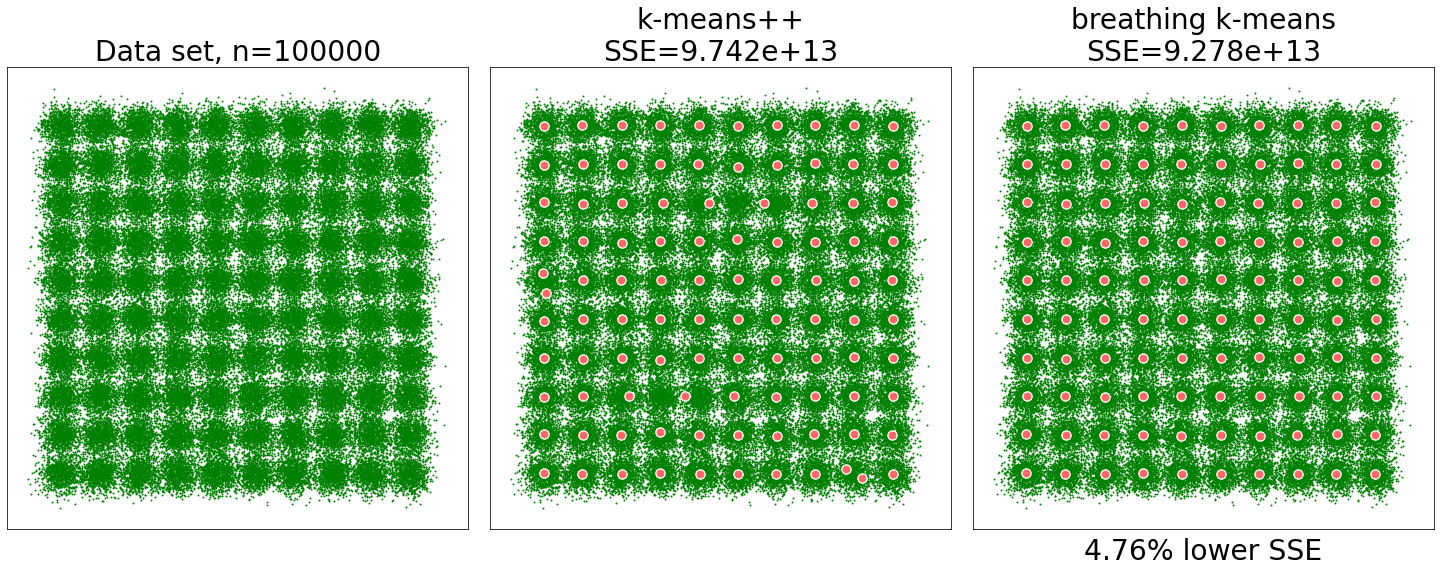
Typical results for the "Birch" data set (100000 points drawn from a mixture of 100 circular Gaussians). k=100
Can you spot the mistakes? :-)
pip install bkmeans
Clone the repository
git clone https://github.com/gittar/breathing-k-meansEnter the top directory.
cd breathing-k-meansCreate the conda environment 'bkm' (or any other name) via
conda env create -n bkm -f environment.ymlActivate the created environment via
conda activate bkmTo run a jupyter notebook with examples, type, e.g.:
jupyter lab notebooks/2D.ipynbThe top level folder contains the following subfolders
-
data/ - data sets used in the notebooks
-
notebooks/ - jupyter notebooks with all examples from the technical report
- 2D.ipynb 2D problems executed with helper functions for brevity
- 2D_detail.ipynb 2D problems executed with raw API
- 10+D.ipynb high-dimensional problems based on the data sets from the original k-means++ publication
-
src/
bkmeans.py- reference implementation of breathing k-means
-
misc/
aux.py- auxiliary functionsdataset.py- general class to administer and plot data setsrunfunctions.py- wrapper functions used in the notebook
The included class BKMeans is subclassed from scikit-learn's KMeans class and has, therefore, the same API. It can be used as a plug-in replacement for scikit-learn's KMeans.
There is one new parameters which can be ignored (left at default) for normal usage:
- m (breathing depth), default: 5
The parameter m can also be used, however, to generate faster ( 1 < m < 5) or better (m>5) solutions. For details see the technical report.
Code:
import numpy as np
from bkmeans import BKMeans
# generate random data set
X=np.random.rand(1000,2)
# create BKMeans instance
bkm = BKMeans(n_clusters=100)
# run the algorithm
bkm.fit(X)
# print SSE (inertia in scikit-learn terms)
print(bkm.inertia_)Output:
1.1775040547902602
Code:
import numpy as np
from sklearn.cluster import KMeans
from bkmeans import BKMeans
# random 2D data set
X=np.random.rand(1000,2)
# number of centroids
k=100
for i in range(5):
# kmeans++
km = KMeans(n_clusters=k)
km.fit(X)
# breathing k-means
bkm = BKMeans(n_clusters=k)
bkm.fit(X)
# relative SSE improvement of bkm over km++
imp = 1 - bkm.inertia_/km.inertia_
print(f"SSE improvement over k-means++: {imp:.2%}")Output:
SSE improvement over k-means++: 3.38%
SSE improvement over k-means++: 4.16%
SSE improvement over k-means++: 6.14%
SSE improvement over k-means++: 6.79%
SSE improvement over k-means++: 4.76%
Kudos go the scikit-learn team for their excellent sklearn.cluster.KMeans class, also to the developers and maintainers of the other packages used: numpy, scipy, matplotlib, jupyterlab