Python API to compute RNA energetics and do structure prediction across multiple secondary structure packages.
Currently supported:
-
Vienna [1] [https://www.tbi.univie.ac.at/RNA/#download]
-
NUPACK [2] [http://www.nupack.org/downloads]
-
RNAstructure [3] [https://rna.urmc.rochester.edu/RNAstructure.html]
-
RNAsoft [4] [http://www.rnasoft.ca/download.html]
-
CONTRAfold [5] [http://contra.stanford.edu/contrafold/]
-
EternaFold [6] [http://https://eternagame.org/about/software]
(c) 2020 Leland Stanford Jr University
Authors: Hannah Wayment-Steele
notebooks: example jupyter notebooks with usage.
scripts: scripts for processing sequences in batch.
parameter_files: dir of various parameter files for packages, put here out of convenience.
test: unit tests (still in work)
mea: code for computing Maximum Expected Accuracy structures.
RNAGraph: DEPRECATED, see https://github.com/DasLab/RiboGraphViz/ for current version of the code. Code to process and visualize secondary structures as graph objects.
- To use Arnie, you will create a file that contains the paths to the software packages that Arnie is wrapping. See
docs/setup_doc.mdfor installation instructions and troubleshooting tips, as well as instructions for setting up the arnie file.
Quickstart: an example file is provided in example_arnie_file.txt.
- Create a variable in your .bashrc:
export ARNIEFILE="/path/to/arnie/<my_file.txt>"
- Add Arnie location to your python path in your .bashrc, i.e.
export PYTHONPATH=$PYTHONPATH:/path/to/arnie
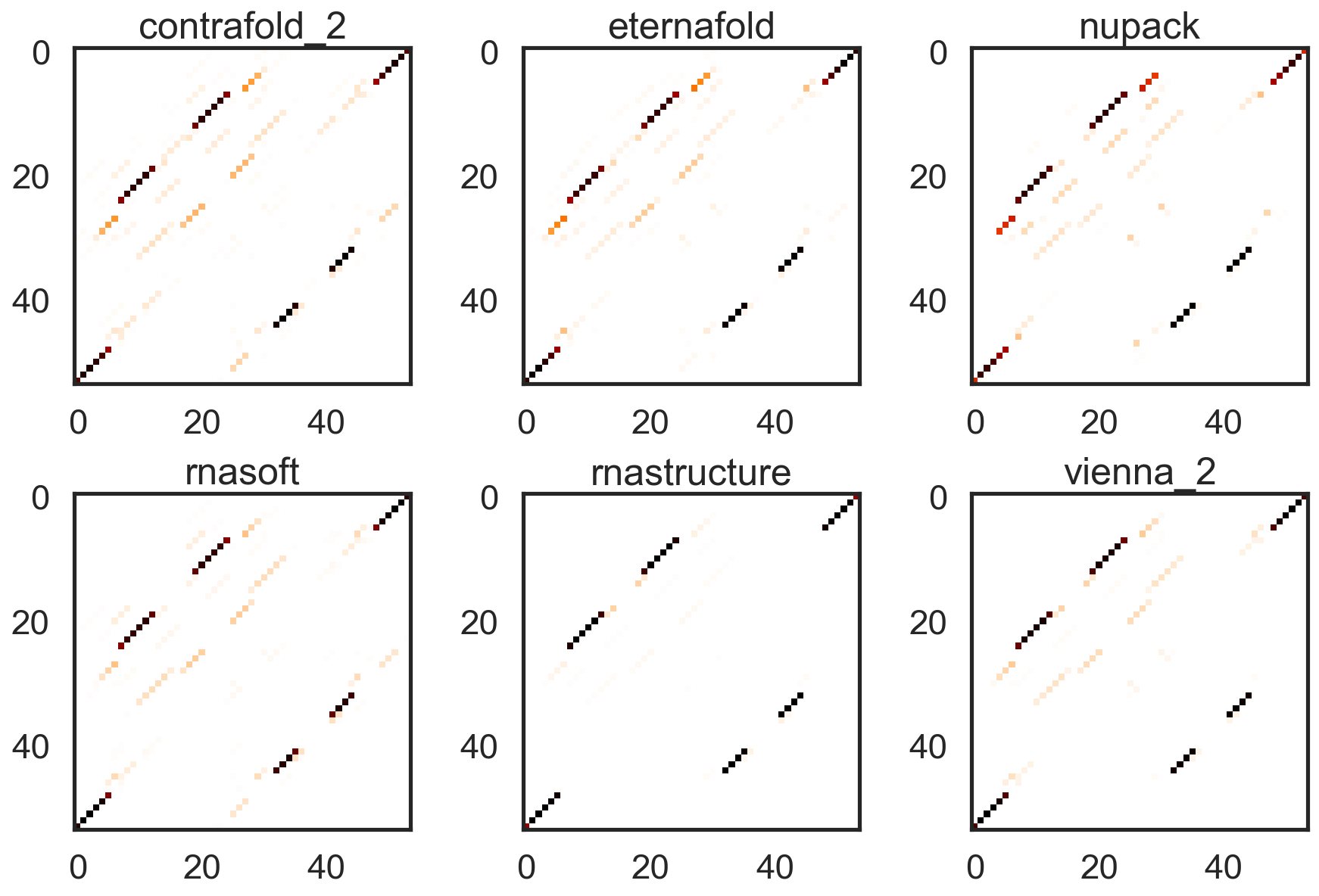
See notebooks/start_here.ipynb for example syntax. In brief, comparing across packages is simple. For computing base pairing probability matrices:
from arnie.bpps import bpps
bpps_dict = {}
my_sequence = 'CGCUGUCUGUACUUGUAUCAGUACACUGACGAGUCCCUAAAGGACGAAACAGCG'
for pkg in ['vienna','nupack','RNAstructure','contrafold','RNAsoft']:
bpps_dict[pkg] = bpps(my_sequence, package=pkg)
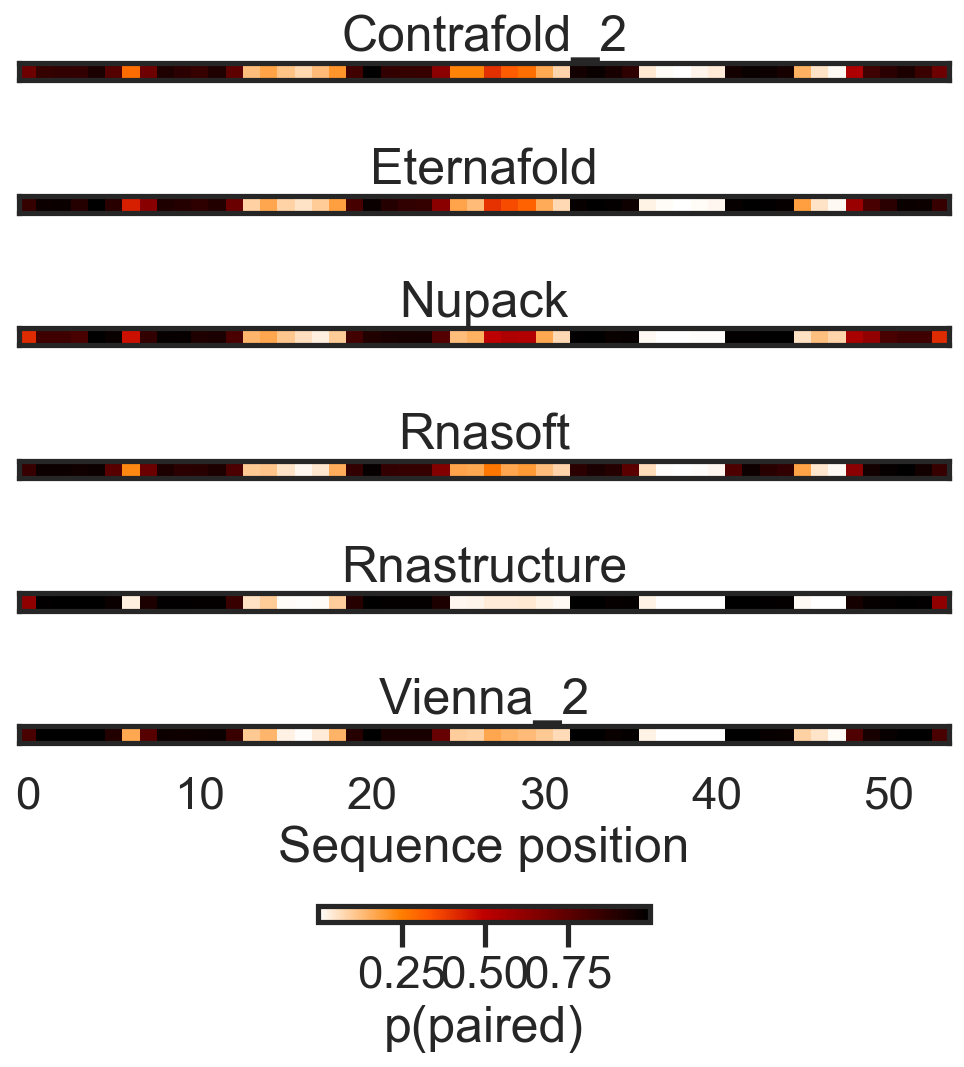
Can also analyze as average base pairing per nucleotide:
- Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol Biol 6, 26 (2011).
- Zadeh, J.N. et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem 32, 170-173 (2011).
- Reuter, J.S. & Mathews, D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129 (2010).
- Andronescu, M., Condon, A., Hoos, H.H., Mathews, D.H. & Murphy, K.P. in RNA, Vol. 16 2304-2318 (2010).
- Do, C.B., Woods, D.A. & Batzoglou, S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 22, e90-98 (2006).
- Wayment-Steele, H.K., Kladwang, W., Eterna Participants, R. Das, Biorxiv (2020).