This repository contains the code for self-supervised pre-training of Swin UNETR model for medical image segmentation. In this readme, you will find a description of Swin UNETR, examples of how to use the code, and links to our datasets and weights.
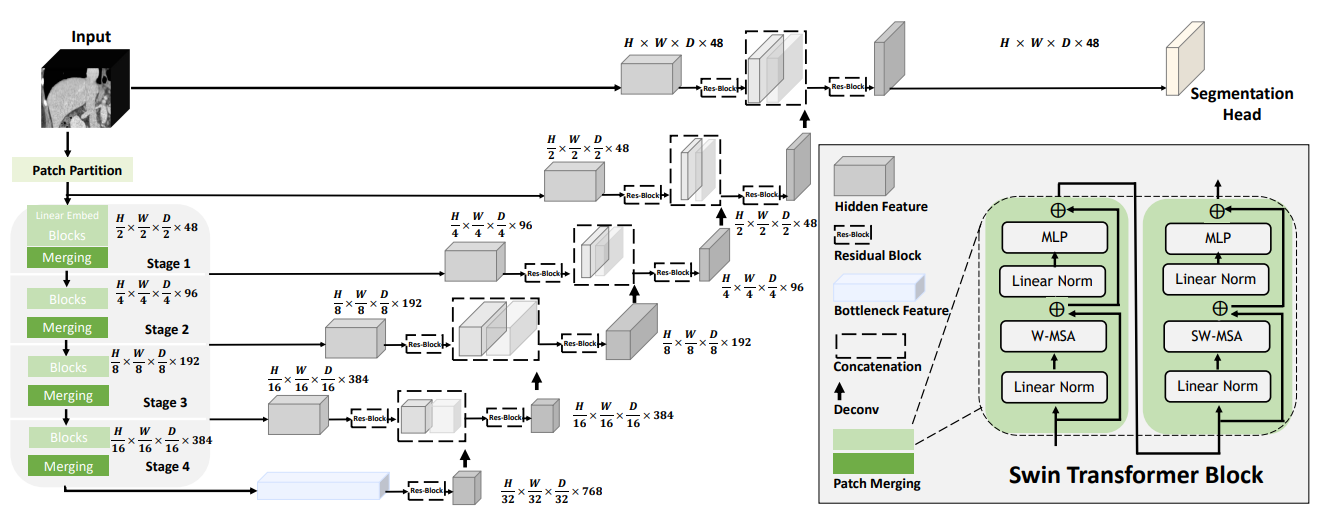
Swin UNETR is the state-of-the-art on Medical Segmentation Decathlon (MSD) and Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset. The architecture of Swin UNETR is illustrated below:
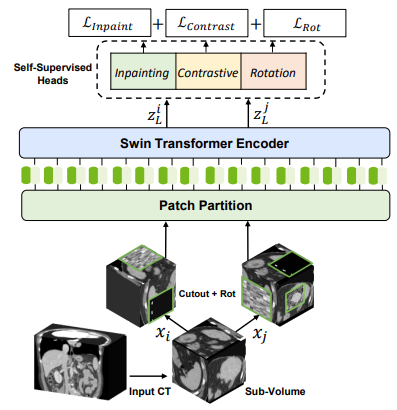
For self-supervised pre-training, randomly cropped tokens are augmented with different transforms such as rotation and cutout and used for pre-text tasks such as masked volume inpainting and contrastive learning and rotation. An overview of the pre-training framework is presented in the following:
The following demonstrates an animation of original images (left) and their reconstructions (right):
Create conda env with yml file and activate
conda env create -f environment.yml
conda activate swin_unetr
We provide the self-supervised pre-trained weights for Swin UNETR backbone (CVPR paper [1]) in this link. In the following, we describe steps for pre-training the model from scratch.
The following datasets were used for pre-training (~5050 3D CT images). Please download the corresponding the json files of each dataset for more details and place them in jsons folder:
- Head & Neck Squamous Cell Carcinoma (HNSCC) (Link) (Download json)
- Lung Nodule Analysis 2016 (LUNA 16) (Link) (Download json)
- TCIA CT Colonography Trial (Link) (Download json)
- TCIA Covid 19 (Link) (Download json)
- TCIA LIDC (Link) (Download json)
To pre-train a Swin UNETR encoder using multi-gpus:
python -m torch.distributed.launch --nproc_per_node=<Num-GPUs> --master_port=11223 main.py
--batch_size=<Batch-Size> --num_steps=<Num-Steps> --lrdecay --eval_num=<Eval-Num> --logdir=<Exp-Num> --lr=<Lr>The following was used to pre-train Swin UNETR on 8 X 32G V100 GPUs:
python -m torch.distributed.launch --nproc_per_node=8 --master_port=11223 main.py
--batch_size=1 --num_steps=100000 --lrdecay --eval_num=500 --lr=6e-6 --decay=0.1To pre-train a Swin UNETR encoder using a single gpu with gradient-checkpointing and a specified patch size:
python main.py --use_checkpoint --batch_size=<Batch-Size> --num_steps=<Num-Steps> --lrdecay
--eval_num=<Eval-Num> --logdir=<Exp-Num> --lr=<Lr> --roi_x=<Roi_x> --roi_y=<Roi_y> --roi_z=<Roi_z>See the LICENSE file for details
If you find this repository useful, please consider citing UNETR paper:
@inproceedings{tang2022self,
title={Self-supervised pre-training of swin transformers for 3d medical image analysis},
author={Tang, Yucheng and Yang, Dong and Li, Wenqi and Roth, Holger R and Landman, Bennett and Xu, Daguang and Nath, Vishwesh and Hatamizadeh, Ali},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
pages={20730--20740},
year={2022}
}
@article{hatamizadeh2022swin,
title={Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images},
author={Hatamizadeh, Ali and Nath, Vishwesh and Tang, Yucheng and Yang, Dong and Roth, Holger and Xu, Daguang},
journal={arXiv preprint arXiv:2201.01266},
year={2022}
}
[1]: Tang, Y., Yang, D., Li, W., Roth, H.R., Landman, B., Xu, D., Nath, V. and Hatamizadeh, A., 2022. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 20730-20740).
[2]: Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H. and Xu, D., 2022. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv preprint arXiv:2201.01266.