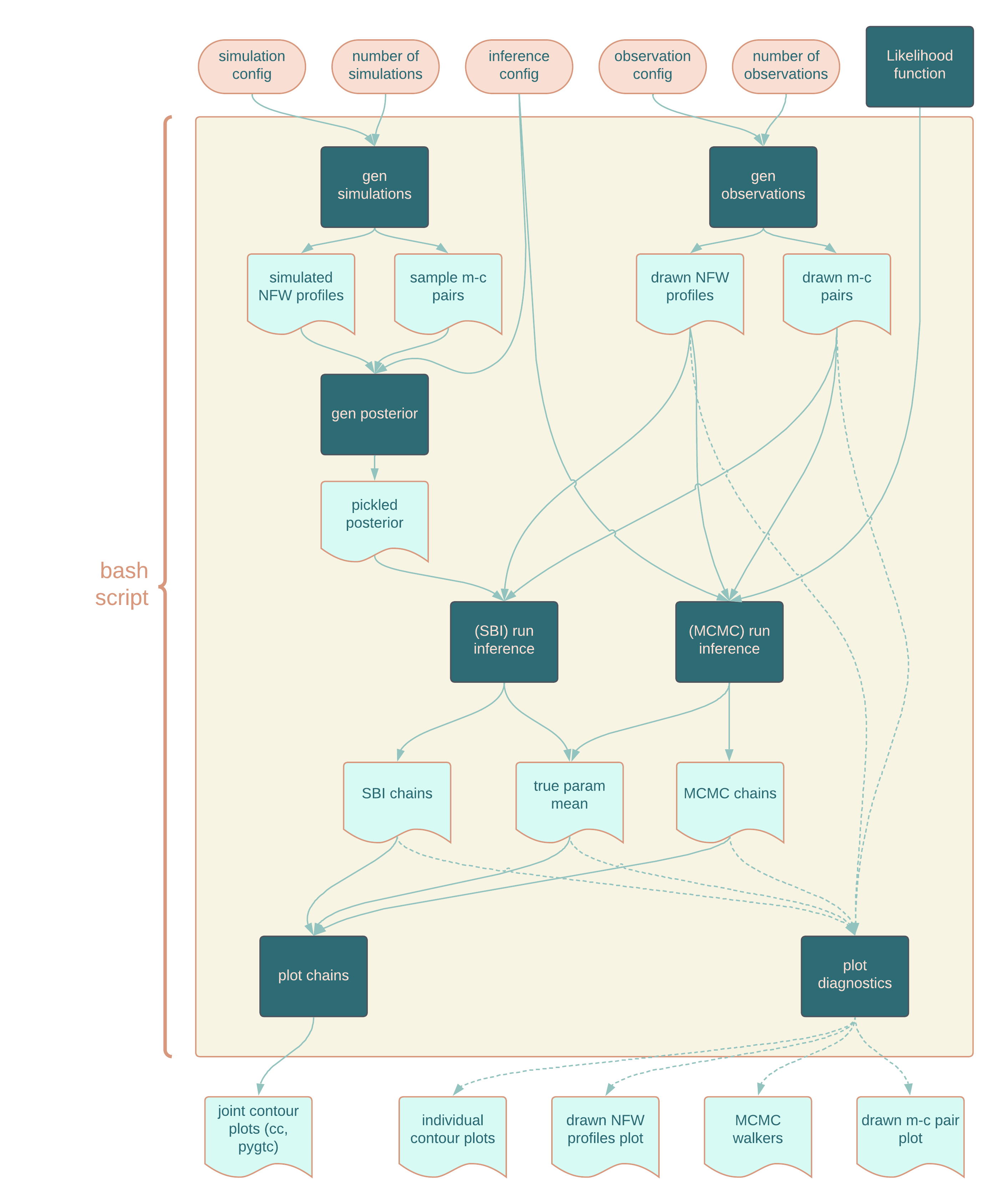
This repository contains tools for experiments for Weak Lensing Galaxy Clusters using Simulation Based Inference.
Summary:
We've split our workflow into four parts:
- Simulation (/configs/simulations) - sim_config tells us about how our simulations were generated/obtained (e.g. the mass-concentration relation used, how much scatter the m-c relation has, the log10mass range, etc).
- Observation(/configs/observations) - obs_config tells us about how our observations were generated/obtained (e.g. the richness-mass relation used, the m-c relation used, the scatter for each of those relations, the richness range, the number of observations, etc).
- Inference (/configs/inference) - infer_config tells us about how our inference was done (e.g. our priors, etc.)
- Plotting - we produce plots using both ChainConsumer and PyGTC.
For each step, the script reading the config will save the output files to the corresponding outputs directory as the config.
Running instructions:
1a) Create a simulation config in the configs/simulations directory
1b) Generate simulations using the following script:
python examples/gen_simulations.py --sim_id {SIM_ID} --num_sims {NUM_SIMS}. This will output to theoutputs/simulations/{SIM_ID}.{NUM_SIMS}directory
2a) Create an inference config in the configs/inference directory
2b) Generate a posterior using the following script:
python examples/gen_posterior.py --sim_id {SIM_ID} --infer_id {INFER_ID} --num_sims {NUM_SIMS}. This will output to theoutputs/posteriors/{SIM_ID}.{INFER_ID}.{NUM_SIMS}directory
3a) Create an observation config in the configs/observations directory
3b) Generate observations using the following script:
python examples/gen_observations.py --obs_id {OBS_ID} --num_obs {NUM_OBS}. This will output to theoutputs/observations/{OBS_ID}.{NUM_OBS}directory
- Run inference using the following script:
python examples/run_inference.py --sim_id {SIM_ID} --infer_id {INFER_ID} --obs_id {OBS_ID} --num_sims {NUM_SIMS} --num_obs {NUM_OBS}. This will output to theoutputs/inference/{SIM_ID}.{INFER_ID}.{OBS_ID}.{NUM_SIMS}.{NUM_OBS}directory
- Plot the contours from the SBI/MCMC chains using the following script:
python examples/plot_chains.py --sim_id {SIM_ID} --infer_id {INFER_ID} --obs_id {OBS_ID} --num_sims {NUM_SIMS} --num_obs {NUM_OBS}. This will output to theoutputs/plots/{SIM_ID}.{INFER_ID}.{OBS_ID}.{NUM_SIMS}.{NUM_OBS}directory