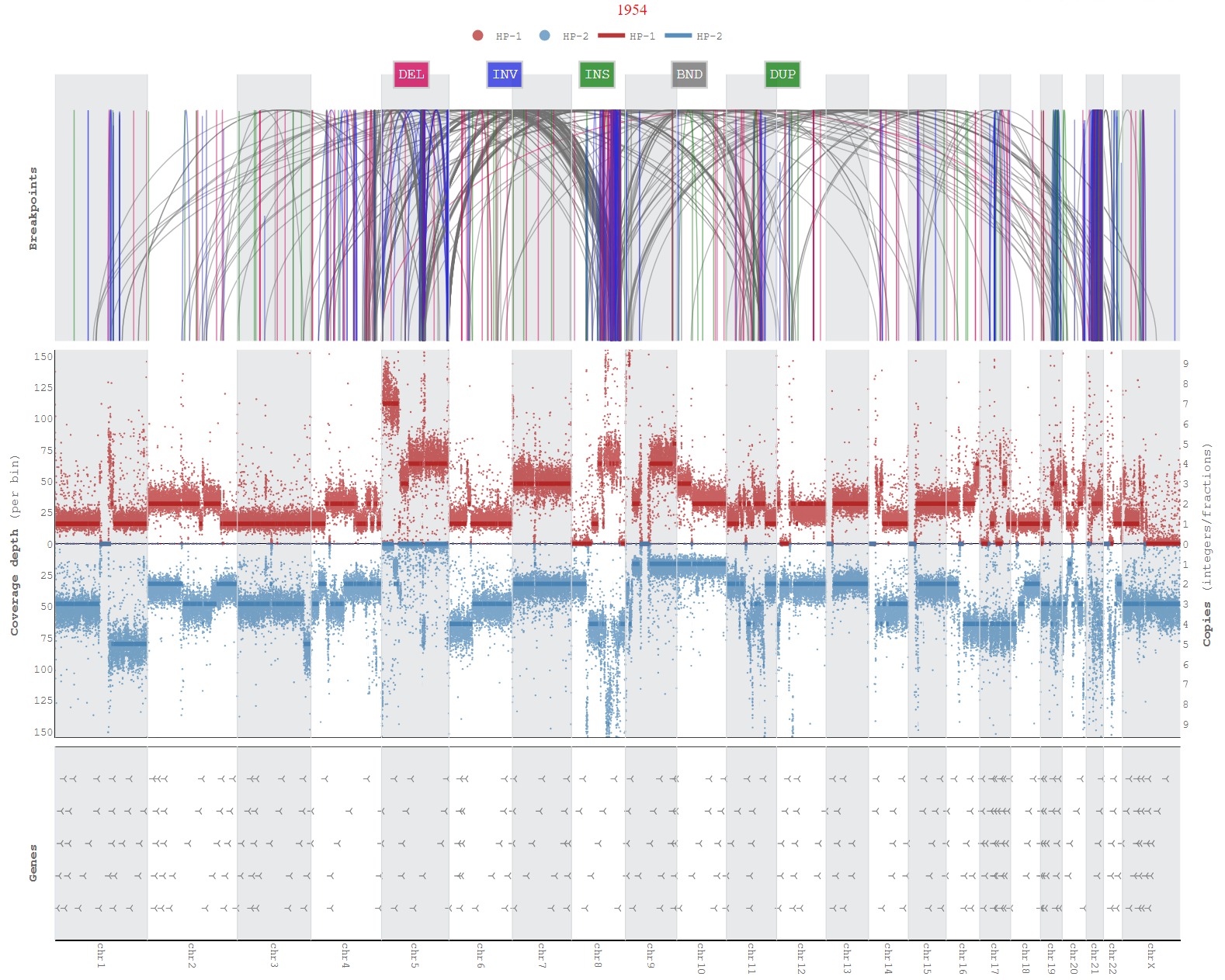
A tool to analyze haplotype-specific chromosome-scale somatic copy number aberrations and aneuploidy using long reads (Oxford Nanopore, PacBio). Wakhan takes long-read alignment and phased heterozygous variants as input, and first uses extends the phased blocks, taking advantage of the CNA differences between the haplotypes. Wakhan then generates inetractive haplotype-specific coverage plots.
git clone https://github.com/KolmogorovLab/Wakhan.git
cd Wakhan/
conda env create -f environment.yml -n Wakhan
conda activate Wakhan
cd src/
python main.py --threads <4> --reference <ref.fa> --target-bam <data.tumor.bam> --normal-phased-vcf <data.normal_phased.vcf.gz> --genome-name <cellline/dataset name> --cut-threshold <150> --out-dir-plots <genome_abc_output> --breakpoints <severus-sv-VCF>
User can input both --tumor-ploidy and --tumor-purity to inform copy number model about normal contamination in tumor to estimate copy number states correctly.
python main.py --threads <4> --reference <ref.fa> --target-bam <data.tumor_haplotagged.bam> --tumor-vcf <data.tumor_phased.vcf.gz> --genome-name <cellline/dataset name> --cut-threshold <150> --out-dir-plots <genome_abc_output> --breakpoints <severus-sv-VCF>
Few cell lines arbitrary phase-switch correction and copy number estimation output with coverage profile is included in the examples directory.
-
--referenceReference file path -
--target-bampath to target bam files (must be indexed) -
--out-dir-plotspath to output coverage plots -
--genome-namegenome cellline/sample name to be displayed on plots -
--normal-phased-vcfnormal phased VCF file to generate het SNPs frequncies pileup for tumor BAM (if tumor-only mode, use phased--tumor-vcfinstead) -
--tumor-vcfVCF file to plot snps frequencies, ratios and LOH regions (Note: phased VCF is required in tumor-only mode) -
--breakpointsFor segmentation to use in CN estimation, structural variations/breakpoints VCF is required
-
--phaseblock-flipping-enableenabling phaseblock flipping in coverage plots -
--phaseblocks-enableenabling phaseblocks display in coverage plots -
--contigsList of contigs (chromosomes, default:chr1-22) to be included in the plots (Note: chrX, chrY support in CNA plots is not included yet) [e.g., chr1-22,X,Y] -
--without-phasingenable it if CNA analysis is being performed without phasing
-
<genome-name>_genome_copynumber.htmlGenome-wide copy number plots with coverage information on same axis -
<genome-name>_copynumber_breakpoints.htmlGenome-wide copy number plots with coverage information on opposite axis, additionally breakpoints and genes annotations -
bed_outputIt contains copy numbers and LOH (in case tumor VCF is provided) segments in bed format -
coverage_plotsHaplotype specific coverage plots for chromosomes with option for unphased coverage -
variation_plotsCopy number chromosomes-scale plots with segmentation, coverage and LOH/SNPs ratios (in case tumor VCF is provided) -
phasing_outputPhase-switch error correction plots and phase corrected VCF file (*rephased.vcf.gz)
This tool requires haplotagged tumor BAM and phased VCF in case tumor-only mode and normal phased VCF in case tumor-normal mode. This can be done through any phasing tools like Margin, Whatshap and Longphase. Following commands could be helpful for phasing VCFs and haplotagging BAMs.
# ClairS phase and haplotag both normal and tumor samples
singularity run clairs_latest.sif /opt/bin/run_clairs --threads 56 --phase_tumor True --use_whatshap_for_final_output_haplotagging --use_whatshap_for_final_output_phasing --tumor_bam_fn normal.bam --normal_bam_fn tumor.bam --ref ref.fasta --output_dir clairS --platform ont_r10
or
# Phase normal sample
pepper_margin_deepvariant call_variant -b normal.bam -f ref.fasta -o pepper/output -t 56 --ont_r9_guppy5_sup -p pepper --phased_output
# Haplotag tumor sample with normal phased VCF (phased.vcf.gz) output from previous step
whatshap haplotag --ignore-read-groups phased.vcf.gz tumor.bam --reference ref.fasta -o tumor_whatshap_haplotagged.bam
# Phase and haplotag tumor sample
singularity run clair3_latest.sif /opt/bin/run_clair3.sh --use_whatshap_for_final_output_haplotagging --use_whatshap_for_final_output_phasing --bam_fn=tumor.bam --ref_fn=ref.fasta --threads=56 --platform=ont --model_path=r941_prom_sup_g5014 --output=clair3 --enable_phasing
or
# Phase and haplotag tumor sample
pepper_margin_deepvariant call_variant -b tumor.bam -f ref.fasta -o pepper/output -t 56 --ont_r9_guppy5_sup -p pepper --phased_output