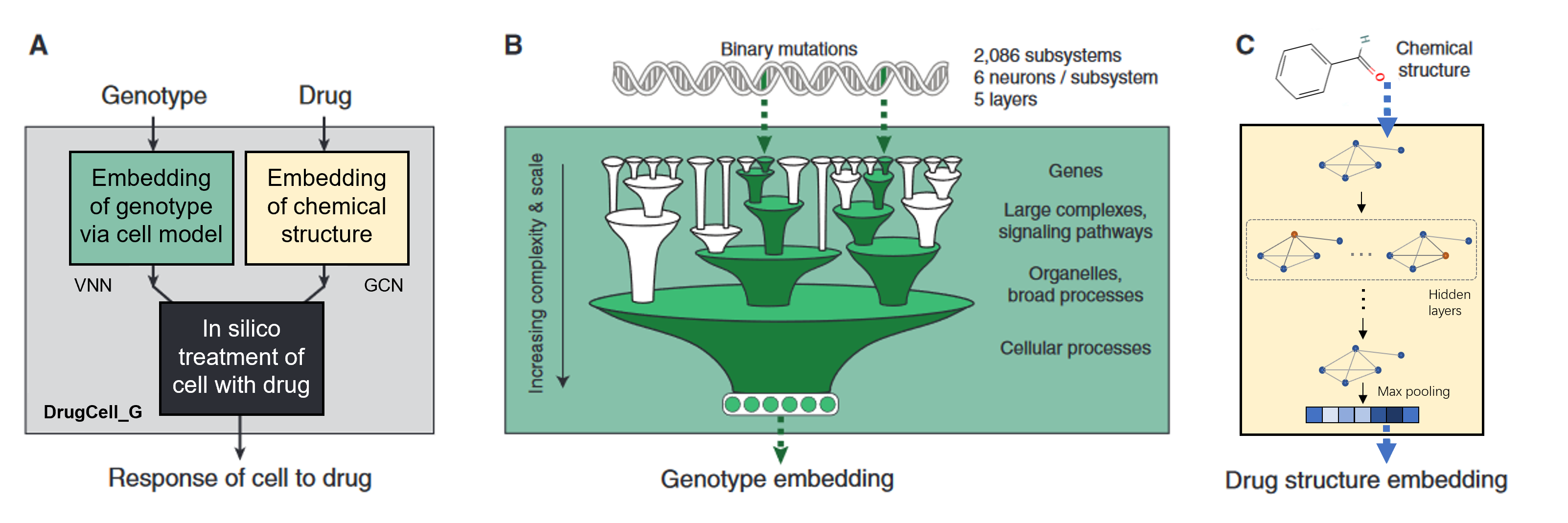
This is the final project of I-529. We did the following experiments based on DrugCell:
- Using unhashed fingerprints of drugs; Comparing the
Baseline1andExp1-1, using hashed or unhashed fingerprint will not effect significantly. - Using Graph Convolution Networks (GCN/GAT) to embed drugs; To traina GCN/GAT in batchwise: We build up the model parallelly shown in following figure, referring this issue. Comparing the
Baseline2andExp2-2, our model performs better in MSE, but not good in PC. More results are coming soon.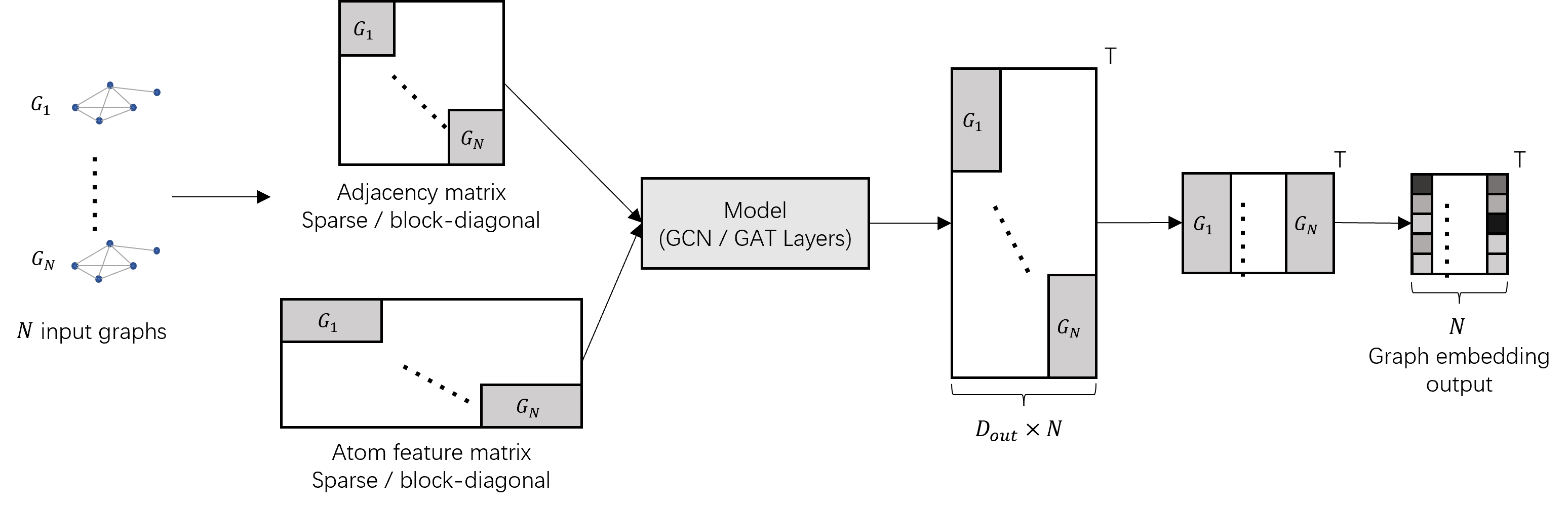
- More metrics: mean mean squared error (MSE), pearson correalation (PC). Different features of these two metrics are here.
The results are:
| Model | Note | PC | MSE | Scripts |
|---|---|---|---|---|
| Baseline0 | Pretrained model* | 0.822805 | 0.014052 | test_pretrain.sh |
| Baseline1 | Train on drugcell_all.txt |
0.828568 | 0.013232 | ours_train.sh & ours_test.sh |
| Exp1-1 | Train on drugcell_all.txt & using unhashed FP |
0.813499 | 0.013995 | ours_train_unhash.sh & ours_test_unhash.sh |
| Exp1-2 | Train on drugcell_all_cut.txt & GCN |
ours_train_gcn.sh & ours_test_gcn.sh |
||
| Exp1-3 | Train on drugcell_all_cut.txt & GAT |
ours_train_gat.sh & ours_test_gat.sh |
||
| Baseline2 | Train on drugcell_train.txt |
0.315630 | 0.282851 | commandline_train.sh & commandline_test_gpu.sh |
| Exp2-2 | Train on drugcell_train.txt & GCN |
-0.036170 | 0.040641 | ours_train_gcn_part.sh & ours_test_gcn_part.sh |
| Exp2-3 | Train on drugcell_train.txt & GAT |
-0.023885 | 0.040629 | ours_train_gat_part.sh & ours_test_gat_part.sh |
The pretrained model can be downloaded here.
The whole dataset can be download here.
$ cat drugcell_all.txt | wc -l
509294
$ cat drugcell_all_cut.txt | wc -l
509280
$ cat drugcell_train.txt | wc -l
10000
$ cat drugcell_test.txt | wc -l
1000Please set up the environment as described in ./DrugCell_README.md. Then install rdkit for loading drug graph and tqdm for showing the process bar by following command:
conda activate pytorch3drugcell
conda install -c rdkit rdkit
conda install -c conda-forge tqdmAll the experiments' scripts are in ./sample/. Please run them as following example:
conda activate pytorch3drugcell
cd sample
# test the pretrained model
./test_pretrain.sh
# train and test our own model
./ours_train.sh
./ours_test.sh
# More experiments' scripts can be found in the table.
./ours_train_unhash.sh
./ours_test_unhash.sh@article{kuenzi2020predicting,
title={Predicting drug response and synergy using a deep learning model of human cancer cells},
author={Kuenzi, Brent M and Park, Jisoo and Fong, Samson H and Sanchez, Kyle S and Lee, John and Kreisberg, Jason F and Ma, Jianzhu and Ideker, Trey},
journal={Cancer cell},
volume={38},
number={5},
pages={672--684},
year={2020},
publisher={Elsevier}
}