nf-core/lasvphylo is a bioinformatics "best-practice" analysis pipeline for creating a high quality alignment of LASV segements and a maximum likelihood tree. I use this pipeline for a fast phylogentic analysis of newly identified LASV cases/outbreaks to determine the rise of new clades or interesting mutations.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It uses Docker/Singularity containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies. Where possible, these processes have been submitted to and installed from nf-core/modules in order to make them available to all nf-core pipelines, and to everyone within the Nextflow community!
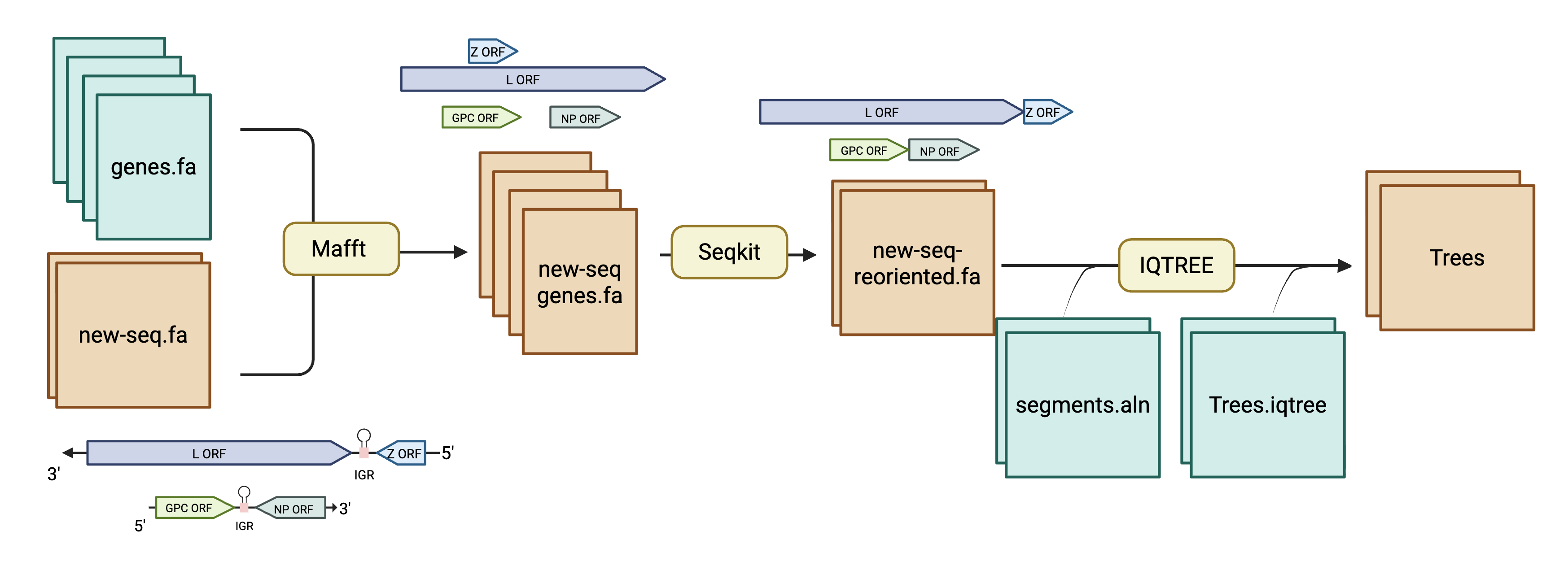
- Orient and isolate the genes of LASV (
MAFFT) - Remove the reference sequences from the alignment (
SeqTk) - Concatenate the genes to each other (
SeqKit) - Align the concatenated genes to an existing alignment (
MUSCLE) - Perform a constrained tree search using a previous tree and new sequences (
IQ-TREE)
-
Install
Nextflow(>=22.10.1) -
Install any of
Docker,Singularity(you can follow this tutorial),Podman,ShifterorCharliecloudfor full pipeline reproducibility (you can useCondaboth to install Nextflow itself and also to manage software within pipelines. Please only use it within pipelines as a last resort; see docs).- The pipeline comes with config profiles called
docker,singularity,podman,shifter,charliecloudandcondawhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker. - If you are using
singularity, please use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs. - If you are using
conda, it is highly recommended to use theNXF_CONDA_CACHEDIRorconda.cacheDirsettings to store the environments in a central location for future pipeline runs.
- The pipeline comes with config profiles called
-
Download the pipeline (git clone) and give in the correct variables :
nextflow run lasvphylo/main.nf -profile YOURPROFILE -c <CONFIG> --outdir <OUTDIR>
Note that some form of configuration will be needed so that Nextflow knows how to fetch the required software. You can chain multiple config profiles in a comma-separated string. In this config file you'll have to specify the following variables:
- input_S : S segment of the new sequence(s)
- input_L : L segment of the new sequence(s)
- input_id_S : id for S segment files
- input_id_L : id for L segment files
- alignment_S : Previous alignment of S segment
- alignment_L : Previous alignment of L segment
- tree_S : Previous tree of S segment
- tree_L : Previous tree of L segment