Dual-scale Vision-Language Multiple Instance Learning for Whole Slide Image Classification, CVPR 2024.
Jiangbo Shi, Chen Li, Tieliang Gong, Yefeng Zheng, Huazhu Fu
[Open Access Version] | [Cite]
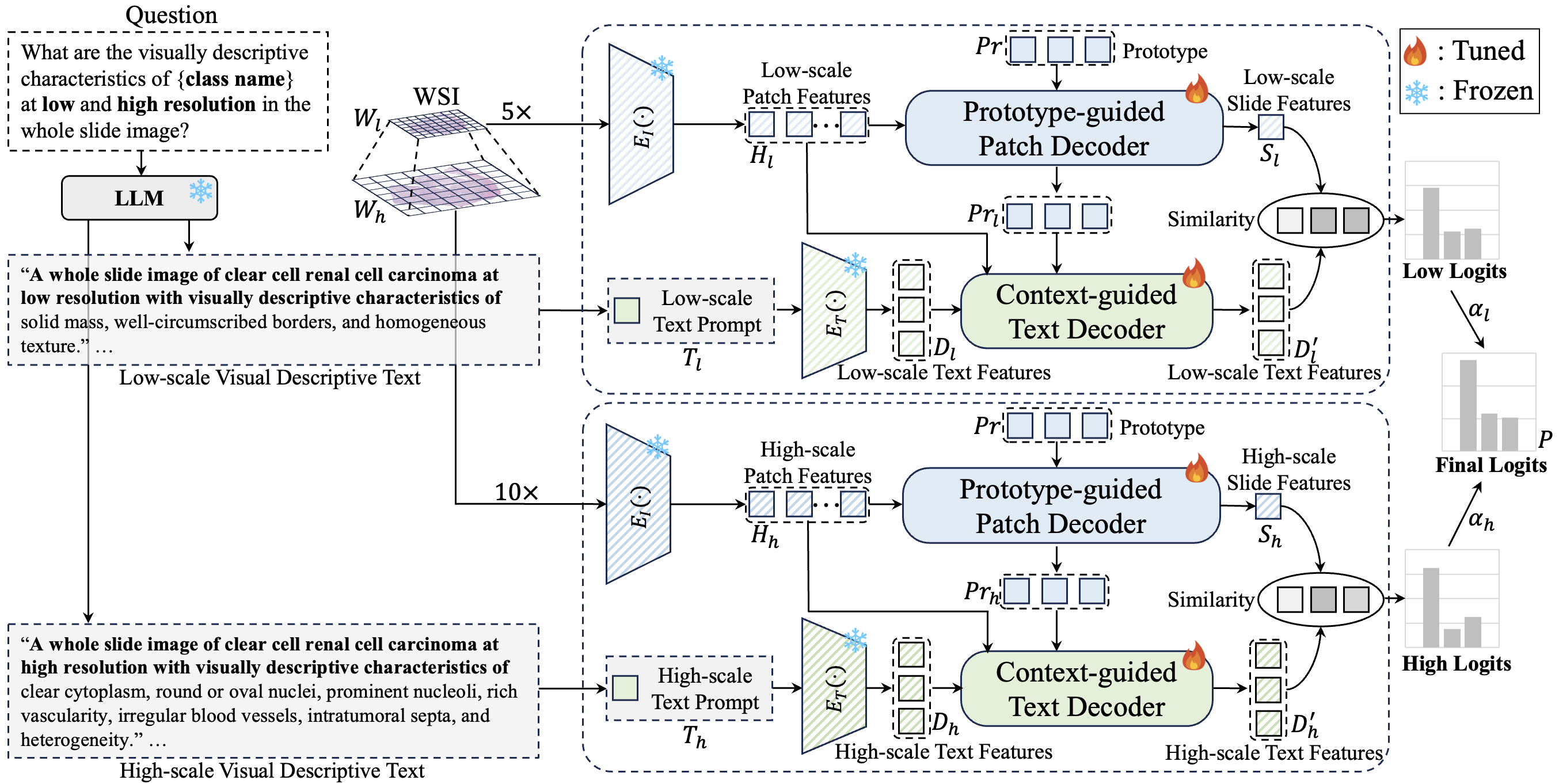
Abstract: Multiple instance learning (MIL)-based framework has become the mainstream for processing the whole slide image (WSI) with giga-pixel size and hierarchical image context in digital pathology. However, these methods heavily depend on a substantial number of bag-level labels and solely learn from the original slides, which are easily affected by variations in data distribution. Recently, vision language model (VLM)-based methods introduced the language prior by pre-training on large-scale pathological image-text pairs. However, the previous text prompt lacks the consideration of pathological prior knowledge, therefore does not substantially boost the model's performance. Moreover, the collection of such pairs and the pre-training process are very time-consuming and source-intensive. To solve the above problems, we propose a dual-scale vision-language multiple instance learning (ViLa-MIL) framework for whole slide image classification. Specifically, we propose a dual-scale visual descriptive text prompt based on the frozen large language model (LLM) to boost the performance of VLM effectively. To transfer the VLM to process WSI efficiently, for the image branch, we propose a prototype-guided patch decoder to aggregate the patch features progressively by grouping similar patches into the same prototype; for the text branch, we introduce a context-guided text decoder to enhance the text features by incorporating the multi-granular image contexts. Extensive studies on three multi-cancer and multi-center subtyping datasets demonstrate the superiority of ViLa-MIL.
Python (3.7.7), h5py (2.10.0), matplotlib (3.1.1), numpy (1.18.1), opencv-python (4.1.1), openslide-python (1.1.1), openslide (3.4.1), pandas (1.1.3), pillow (7.0.0), PyTorch (1.6.0), scikit-learn (0.22.1), scipy (1.4.1), tensorboardx (1.9), torchvision (0.7.0), captum (0.2.0), shap (0.35.0), clip (1.0).
The two public TCGA-RCC and TCGA-Lung datasets can be downloaded in NIH Genomic Data Commons Data Portal. For the specific downloading tool, please refer to GDC Data Transfer.
For each dataset, a .csv file is needed in the following format and put it into the dataset_csv folder:
Headname: 'case_id, slide_id, label'
Each line: 'patient_0, TCGA-UZ-A9PS-01Z-00-DX1.3CAF5087-CDAB-42B5-BE7C-3D5643AEAA6D, CCRCC'
4.1 Generate Patch Coordinate
Utilize the sliding window method to generate the patch coordinates from WSI for cropping.
python create_patches_fp.py \
--source SOURCE_FOLDER \
--slide_name_file SLIDE_NAME_FILE \
--preset tcga.csv \
--save_dir SAVE_FOLDER \
--patch_size 1024 \
--step_size 1024 \
--seg \
--patch \
--uuid_name_file UUID_NAME_FILEThe list of parameters is as follows:
source: The downloading original WSI folder path instep 2.slide_name_file: The path of csv dataset file instep 3.save_dir: The saving folder path for generating results.uuid_name_file: A file stores the corresponding relations between uuid and slide name in the followling format ('uuid', 'slide name', 'slide label').
4.2 Crop Patches
Utilize the above generated patch coordinate file to crop the patches.
cd feature_extraction
python patch_generation.py
Note: the pathes between line 12 and 17 are needed to define in patch_generation.py.
Paramters Descriptions:
slide_folder: The downloading original WSI folder path instep 2.all_data: The same path as theuuid_name_fileinstep 4.1.root_folder: The same path as the save_dir instep 4.1.define_path_size: For the TCGA dataset, all the patches are cropped at the highest magnification. If patches are needed to crop at 10x with size 256. this parameter should defined as 1024.save_folder: The path where the cropping patches are saved.
4.3 Extract Patch Features
Utilize a pretrain encoder (e.g., CLIP-pretrained ResNet50) to extract the patch features.
python patch_extraction.py \
--patches_path 'PATCHES_PATH' \
--library_path 'LIBRARY_PATH' \
--model_name 'clip_RN50' \
--batch_size 64 \
Parameter Descriptions:
patches_path: The same as the save_folder instep 4.2.libiary_path: The path where the generated patch features are saved.model_name:'clip_RN50'means using CLIP ResNet50 to extract patch features.'resnet50_trunc'means usingResNet50pretrained on ImageNet to extrach features.
5.1 Generate k splitting datasets with different seeds.
python create_splits_seq.py \
--label_frac 1 \
--k 5 \
--task 'TASK' \
--val_frac VAL_FRAC \
--test_frac TEST_FRAC \
--dataset DATASET \
Parameter Descriptions:
task: 'task_tcga_rcc_subtyping' for the TCGA_RCC dataset.val_frac: The proportion of the validation set (e.g., 0.3).test_frac: The proportion of the test set (e.g., 0.3).dataset: Dataset name (e.g., TCGA_RCC).
5.2 Build the few-shot dataset split.
python create_splits_fewshot.py
Note: the parameters are needed to define between the line 5 and 8 in the create_splits_fewshot.py.
Parameter Descriptions:
N: The case number in training set for each category.data_foler: The path generated instep 5.1.all_data: The same path asuuid_name_fileinstep 4.1.save_folder: The saved split file path.
By querying the frozen LLM (e.g., GPT-3.5) with the following question, the text prompt will be generated automatically.
Q: What are the visually descriptive characteristics of {class name} at low and high resolution in the whole slide image?
The specific text prompt files for TCGA-RCC and TCGA-Lung datasets are stored in the folder text_prompt using GPT-3.5.
Run the following script, and ViLa-MIL will be trained.
export CUDA_VISIBLE_DEVICES=0
python main.py \
--seed 1 \
--drop_out \
--early_stopping \
--lr 1e-4 \
--k 5 \
--label_frac 1 \
--bag_loss ce \
--task 'TASK' \
--results_dir 'RESULT_DIR' \
--exp_code 'EXP_CODE' \
--model_type ViLa_MIL \
--mode transformer \
--log_data \
--data_root_dir 'DATA_ROOT_DIR' \
--data_folder_s 'DATA_FOLDER_S' \
--data_folder_l 'DATA_FOLDER_L' \
--split_dir 'SPLIT_DIR' \
--text_prompt_path 'TEXT_PROMPT_PATH' \
--prototype_number 16 \
Parameter Descriptions:
task: 'task_tcga_rcc_subtyping' for the TCGA_RCC dataset.results_dir: The path where model training results are saved.exp_code: The folder saved inresults_dir.data_root_dir: The path where patch features are saved.data_folder_s: The folder name for low-scale patch features.data_folder_l: The folder name for high-scale patch features.split_dir: The same path assave_folderinstep 5.2.text_prompt_path: The path of text prompt file.
Eval the trained model using the following script.
export CUDA_VISIBLE_DEVICES=0
python eval.py \
--drop_out \
--k 5 \
--k_start 0 \
--k_end 5 \
--task 'TASK' \
--results_dir 'RESULTS_DIR' \
--models_exp_code 'MODELS_EXP_CODE' \
--save_exp_code 'SAVE_EXP_CODE' \
--model_type ViLa_MIL \
--mode transformer \
--splits_dir 'SPLITS_DIR' \
--data_root_dir 'DATA_ROOT_DIR' \
--data_folder_s 'DATA_FOLDER_S' \
--data_folder_l 'DATA_FOLDER_L' \
--text_prompt_path 'TEXT_PROMPT_PATH' \
--prototype_number 16 \
Parameter Descriptions:
task: 'task_tcga_rcc_subtyping' for TCGA_RCC dataset.results_dir: The path where model training results are saved.models_exp_code: The training model saving path.save_exp_code: The path of saving the evaluation results.split_dir: The same path assave_folderinstep 5.2.data_root_dir: The path where patch features are saved.data_folder_s: The folder name for low-scale patch features.data_folder_l: The folder name for high-scale patch features.text_prompt_path: The path of the text prompt file.
Note that the symboles
If you find our work useful in your research, please consider citing our paper at:
@inproceedings{shi2024vila,
title={{ViLa-MIL}: Dual-scale Vision-Language Multiple Instance Learning for Whole Slide Image Classification},
author={Shi, Jiangbo and Li, Chen and Gong, Tieliang and Zheng, Yefeng and Fu, Huazhu},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
pages={11248--11258},
year={2024}
}
This project is based on CLAM, CoOp, and Self-Supervised-ViT-path. We have great thanks for these awesome projects.