|\ \ \ \ \ \ \ \ __ ___
| \ \ \ \ \ \ \ \ | O~-_ _-~~ ~~-_
| >----|-|-|-|-|-|-|--| __/ / BELIEVE )
| / / / / / / / / |__\ < )
|/ / / / / / / / \_ ME ! _)
~--___--~
Associations search is one of the methods of data analysis. Associated Rule Mining (ARM) approach can construct association rules from observational data, but the most widely used algorithm Apriori typically produces a large number of unstructured results without any ranking or statistical significance. We propose a novel method FARM (Fishbone Association Rule Mining) to address these challenges. FARM is a method for hierarchical association rule construction with significance filtering and a web-application for interactive visualisation and exploration of results. Hierarchical rules are visualized in a form of Fishbone Ishikawa diagrams. Fishbone is a client-server application for discovering and visualization of hierarchical rules with interactive web UI and http API.
Open Access Paper: https://doi.org/10.1145/3459930.3469499
Citation: Tsurinov, P., Shpynov, O., Lukashina, N., Likholetova, D. and Artyomov, M. FARM: hierarchical association
rule mining and visualization method.
In Proceedings of the 12th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (pp. 1-1),
2021
Poster is available here.
This service implements a novel approach to mine association rules within specified data.
In addition to constructing hierarchical rules, it implements filtering of unproductive rules according to 'improvement'
metric [1] with corresponding significance check.
Significance check is done using holdout approach [2].
- Java 11
- Gradle >= 5.51
Clone bioinf-commons library under the project root.
git clone git@github.com:JetBrains-Research/bioinf-commons.git
Launch the following command line to build jar:
./gradlew shadowJar
This command will create executable jar file with name
build/libs/fishbone-{version}.build.jar
In order to test service, use the following command from the project directory:
./gradlew test
This command will run all available kotlin tests.
To run service use the following command:
java -Dgenomes.path={path_to_empty_folder_to_store_genomes} -jar build/libs/fishbone-{version}.build.jar
Optional program options are:
--port{port to run server; default: 8080}--output{path to output folder}
Web UI consists of two main parts: running analysis and visualize results from local files.
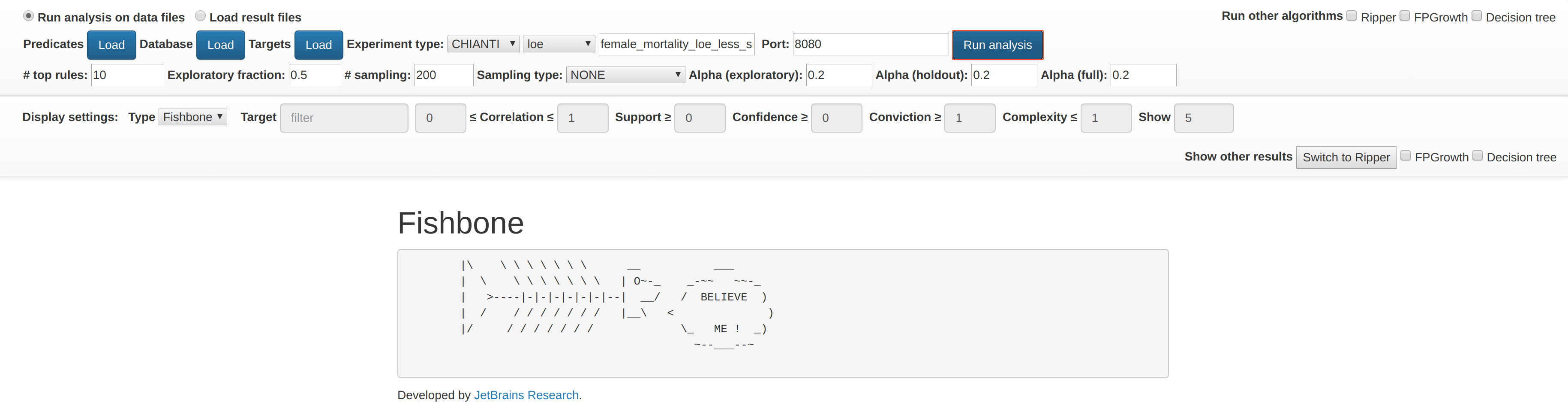 Predicate files, database file, experiment type and objective function must be specified to run Fishbone algorithm.
Checkboxes on the right side could be also used to run alternative algorithms.
Predicate files, database file, experiment type and objective function must be specified to run Fishbone algorithm.
Checkboxes on the right side could be also used to run alternative algorithms.
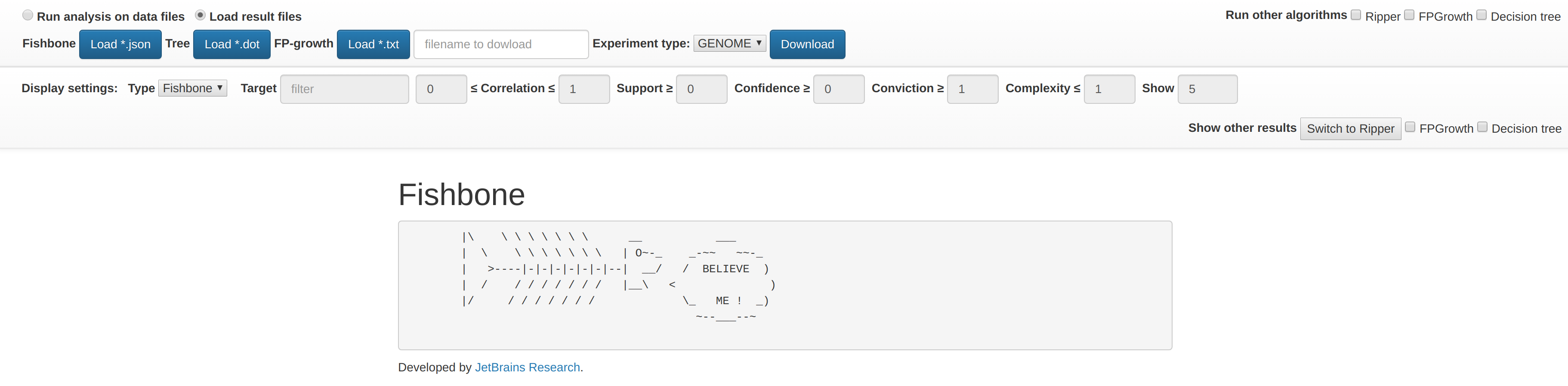 Local result files for different algorithms could be selected to visualize.
Local result files for different algorithms could be selected to visualize.