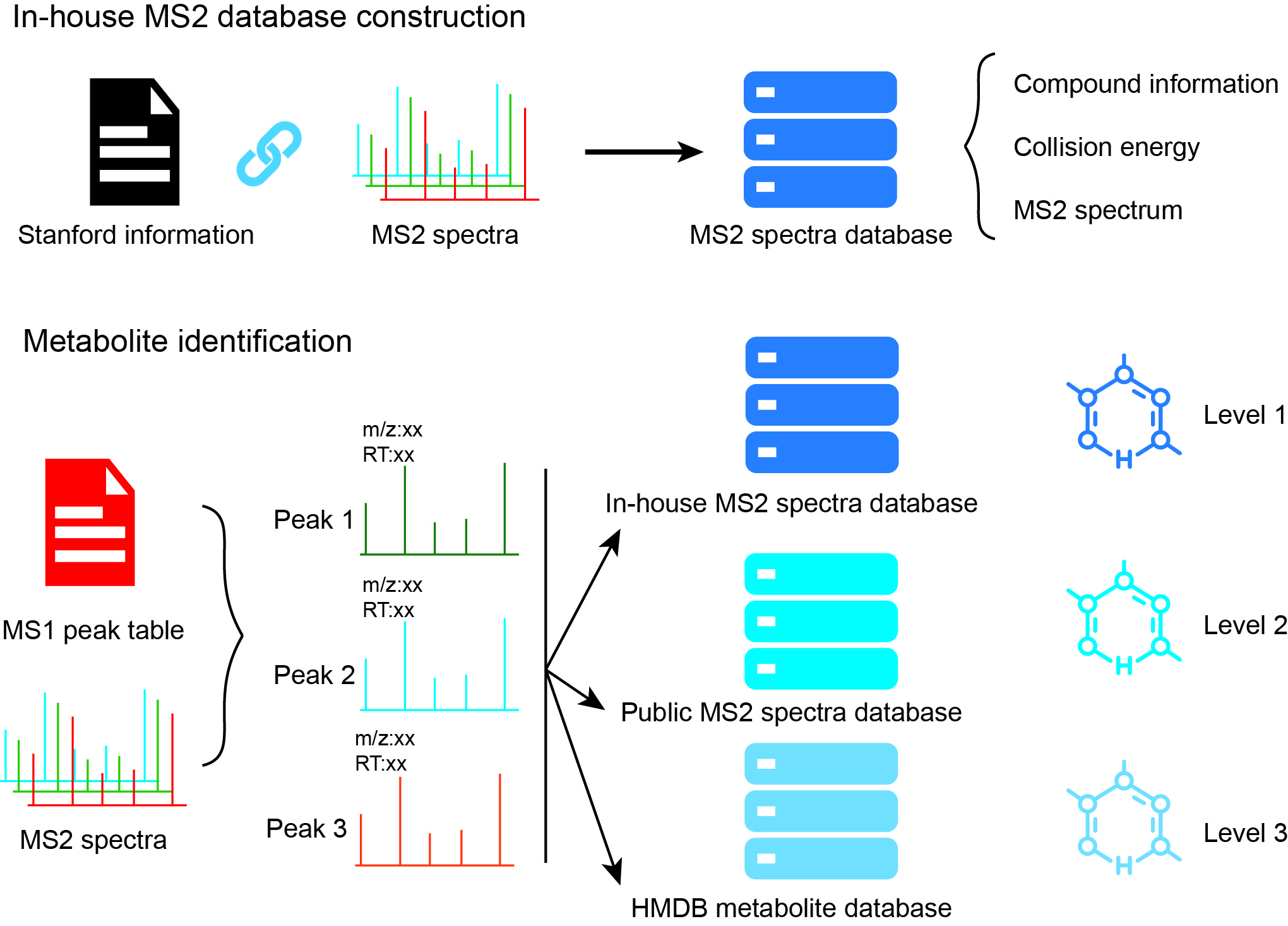
metID is a R packge which is used for metabolite identification based
on in-house database and public database based on accurate mass (m/z),
rentention time (RT) and/or MS2
spectra.
You can install metID from
Github.
if(!require(devtools)){
install.packages("devtools")
}
devtools::install_github("jaspershen/metID")tinyTools is a dependent package for metID. Please install it too.
if(!require(devtools)){
install.packages("devtools")
}
devtools::install_github("jaspershen/tinyTools")Please see the Get
started page to
get the instruction of metID.
If you have any quesitions about metID, please don’t hesitate to email
me (shenxt@stanford.edu).
M339, Alway Buidling, Cooper Lane, Palo Alto, CA 94304
If you use metID in you publication, please cite this publication:
X. Shen, R. Wang, X. Xiong, Y. Yin, Y. Cai, Z. Ma, N. Liu, and Z.-J.
Zhu* (Corresponding Author), Metabolic Reaction Network-based Recursive
Metabolite Annotation for Untargeted Metabolomics, Nature
Communications, 2019, 10: 1516.
Web Link.
Thanks very much!