An open-source Python tool for high-throughput chemistry.
PyPlate provides tools for the design and implementation of high-throughput chemistry experiments (in particular, reaction screening). It allows the user to define a space of experimental parameters to be explored, select points in that space for experimentation, and design liquid/solid handling steps to implement those experiments in 96 well plates.
PyPlate requires Python 3.10 or later.
pip install pyplate-hte
To view plate visualizations, you will need an interactive Python shell like Jupyter.
All experiments are divided into a design phase and an implementation phase.
Design Phase: TBD
Implementation Phase: PyPlate mimics the physical process of dispensing solids or liquids into plates. Substances are placed into Containers and dispensed into Plates. The instructions for creating a set of plates are grouped into Recipe objects.
- add something about how to check that the implementation and design are consistent
- All objects in PyPlate are considered immutable.
from pyplate import Substance, Container, Plate, Recipe
triethylamine = Substance.liquid(name="triethylamine", mol_weight=101.19, density=0.726)
DMSO = Substance.liquid(name="DMSO", mol_weight=78.13, density=1.1004)
triethylamine_50mM = Container.create_solution(solute=triethylamine, solvent=DMSO, concentration='50 mM', total_quantity='10 mL')
plate = Plate(name='plate', max_volume_per_well='50 uL')
recipe = Recipe().uses(triethylamine_50mM, plate)
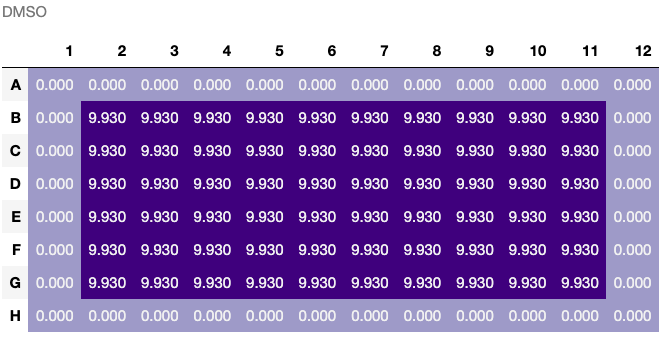
recipe.transfer(source=triethylamine_50mM, destination=plate[2:7, 2:11], quantity='10 uL')
triethylamine_50mM, plate = recipe.bake()
recipe.steps[-1].visualize(what='destination', mode='final', unit='uL')[0]A Substance is a solid, liquid, or enzyme:
# solid
sodium_chloride = Substance.solid(name="Sodium Chloride", mol_weight=58.44)
# liquid
water = Substance.liquid(name="Water", mol_weight=18.01528, density=1.0)
# enzyme
enzyme_X = Substance.enzyme(name="Enzyme X")Enzymes are like solids, but are specified in units of activity instead of moles. The default density for substances is 1.0.
A Container holds defined amounts of a set of Substances and has a maximum volume.
Placing 10 g of NaCl in a 1 L container:
sodium_chloride_stock = Container(name = "sodium_chloride_stock",
max_volume = "1 L",
initial_contents =
[(sodium_chloride, "10 g")])Making 10 mL of a 0.01 M stock solution of NaCl in water:
sodium_chloride_10mM = Container.create_solution(
solute = sodium_chloride,
solvent = water,
concentration = "0.01 M",
total_quantity = "10 mL")Diluting the above solute to 0.005 M:
sodium_chloride_10mM, sodium_chloride_5mM = Container.create_solution_from(source=sodium_chloride_10mM, solute=sodium_chloride, concentration="5 mM", solvent=water, quantity="10 mL")Containers can be diluted to a desired concentration or filled to a desired volume.
salt_water = salt_water.dilute(solute=sodium_chloride_10mM, concentration='5 mM', solvent=water)
salt_water = salt_water.fill_to(solvent=water, quantity='50 mL')Plates are rectangular arrays of Containers.
To create a generic 96 well plate:
plate = Plate(name="test plate", max_volume_per_well="50 uL")
Custom plates can be created with different numbers of rows and columns as well as different labels:
plate = Plate(name="custom plate", max_volume_per_well="50 uL", rows=16, columns=24)
plate = Plate(name="custom plate", max_volume_per_well="50 uL", rows=['i', 'ii', 'iii'], columns=['a', 'b', c'])
PyPlate follows the pandas convention of having both integer- and label-based indices for referencing wells in Plates. When row or column specifiers are provided as integers, they are assumed to be integer indices (1, 2, 3, ...). When specifiers are provided as strings, they are assumed to be label indices ("A", "B", "C", ...).
By default, rows in plates are given alphabetical labels "A", "B", "C", ... and columns in plates are given numerical labels 1, 2, 3. However, rows and columns are always given integer indices 1, 2, 3, .... For example, "B:3" and (2,3) both refer to well B3.
Here are some ways to refer to a specific well:
- String Method: "A:1"
- Tuple Method: ('A', 1)
You can refer to multiple wells as a list:
plate[[('A', 1), ('B', 2), ('C', 3), 'D:4']]
Slicing syntax is supported:
- In addition, you can provide python slices of wells with 1-based indexes:
plate[:3], plate[:, :3], plate['C':], plate[1, '3':]
A Recipe is a set of instructions for transforming one set of containers into another.
plate = Plate(name='plate', max_volume_per_well='50 uL')
recipe = Recipe()Containers and Plates must be declared in the Recipe before use:
recipe.uses(plate)It can be convenient to create solutions and declare them for use in the same step:
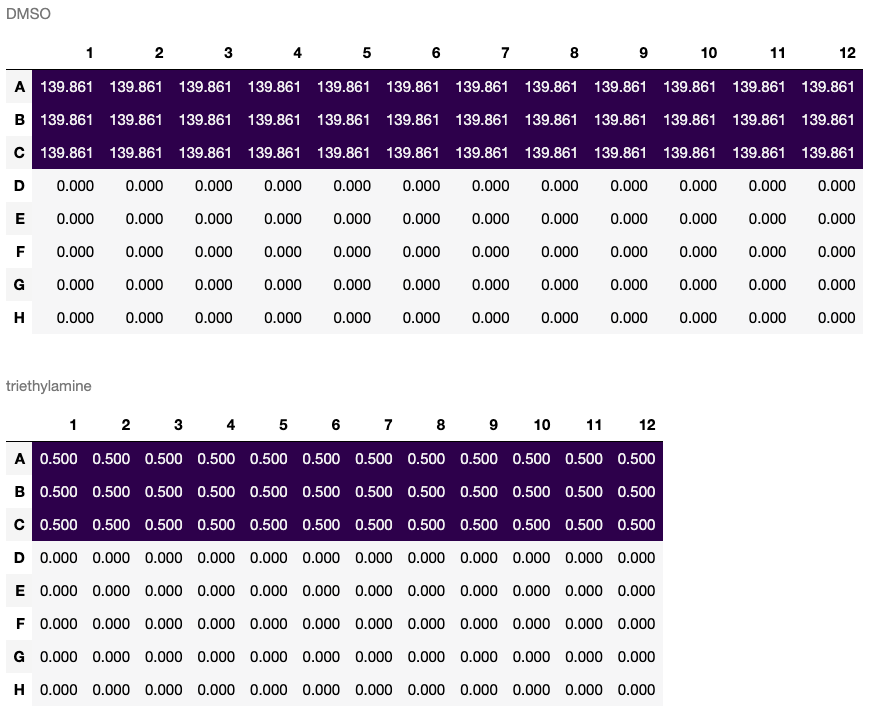
triethylamine_50mM = recipe.create_solution(solute=triethylamine, solvent=DMSO, concentration='0.05 M', total_quantity='10.0 mL')(Note that solutions made in this way are not actually created until recipe.bake() is called.) Performing transfer steps:
recipe.transfer(source=triethylamine_50mM, destination=plate[:3], quantity='10 uL')When recipe.bake() is called, the resulting Container and Plate objects are returned (leaving the input objects, if any, unchanged) in order of declaration or creation:
plate, triethylamine_50mM = recipe.bake()
Each operation called on a recipe is stored as a step. You can retrieve the steps for a recipe using recipe.steps.
Each step has instructions as to what happened during the step (step.instruction) as well as visualizations.
Each step in a recipe that involves a plate will yield visualizations. We can visualize information about the source or destination plate (as applicable) or both. Visualization can be determined for two different modes, either the 'final' state of the plate or a 'delta' of what changed during the step. Finally, the unit in which data is returned must be provided.
...
last_step = recipe.steps[-1]
for df in last_step.visualize(what='destination', mode='delta', unit='umol'):
display.display_html(df)
The basic units of pyplate are moles, grams, liters, and activity units. ('mol', 'g', 'L', 'U')
Any time units are required, metric prefixes may be specified. ('mg', 'umol', 'dL', ...)
All quantities are specified as strings with a value and a unit. ('1 mmol', '10 g', '10 uL' ...)
Documentation can be built by executing make docs. The resulting documentation will be in the docs folder.
Tests can be run by executing pytest.
Licensed under the Apache License, Version 2.0 (the "License") You may obtain a copy of the License at https://www.apache.org/licenses/LICENSE-2.0