MutaFrame is a single page web app that makes diease predictions on Human Proteome accessible.
We provide client side ui at mutaframe.com and also several other APIs with proper Access-Control-Allow-Origin: * headers . When you use our apis, your base uri is always https://deogen2.mutaframe.com/. You need to make a [GET] request to the appropriate path to receive a response.
General rate limiting rules apply, you can make up to ~45 calls per minute, which after you'll start receiving 403. After a minute, you should be good to go again.
You can search for a gene symbol, accession or a phrase enclosed between quotes, you receive a json response of the information available.
| field | desc | default | required |
|---|---|---|---|
| query | alpha numeric | NA | required |
| species | common name or taxonomy id | human | optional |
| isoforms | retrieve all isoforms | false | optional |
| refseq | retrieve only refseq | false | optional |
Array
GET https://deogen2.mutaframe.com/tools/pseudo?query="breast cancer 2"
[{"mrna":"NM_000059.4","protein":"NP_000050.3","uniprot":"P51587","name":"BRCA2 DNA repair associated","symbol":"BRCA2"}]
GET https://deogen2.mutaframe.com/tools/pseudo?query=TUBA1A&isoforms=true
[{"mrna":"NM_001270399.1","protein":"NP_001257328.1","uniprot":"Q71U36","name":"tubulin alpha 1a","symbol":"TUBA1A"},{"mrna":"NM_001270400.1","protein":"NP_001257329.1","uniprot":"Q71U36","name":"tubulin alpha 1a","symbol":"TUBA1A"},{"mrna":"NM_006009.4","protein":"NP_006000.2","uniprot":"Q71U36","name":"tubulin alpha 1a","symbol":"TUBA1A"}]
GET https://deogen2.mutaframe.com/tools/pseudo?query=NM_001130916.2&refseq=true
[{"mrna":"NM_004612.4","protein":"NP_004603.1","uniprot":"P36897","name":"transforming growth factor beta receptor 1","symbol":"TGFBR1"}]
Provide an accession number and receive back the sequences if the latest refseq version exists.
| field | desc | default | required |
|---|---|---|---|
| accession | entrez accession | NA | required |
| fields | protein,mRNA | NA | required |
| response | return format | json | optional |
| download | trigger download | false | optional |
| refseq | only refseq | false | optional |
Object
GET https://deogen2.mutaframe.com/tools/seqfetch?accession=NM_001130916.2&fields=mrna,protein&response=json
//{"protein":"MEAAVAAP...","mrna":"aaagggccggagcga..."}
GET https://deogen2.mutaframe.com/tools/seqfetch?accession=NM_001130916.2&fields=protein&response=json
//{"protein":"MEAAVAAP..."}
GET https://deogen2.mutaframe.com/tools/seqfetch?accession=NM_001130916.2&fields=protein&response=fasta
//> NM_001130916.2 protein
MEAAVAAPRPRLL...
You can trigger a browser side download as well:
GET https://deogen2.mutaframe.com/tools/seqfetch?accession=NM_001130916.2&fields=protein&response=json&download=true
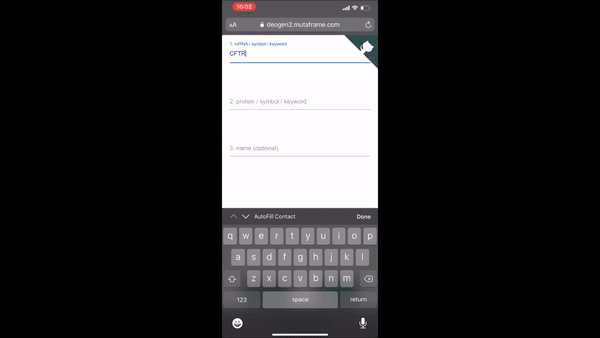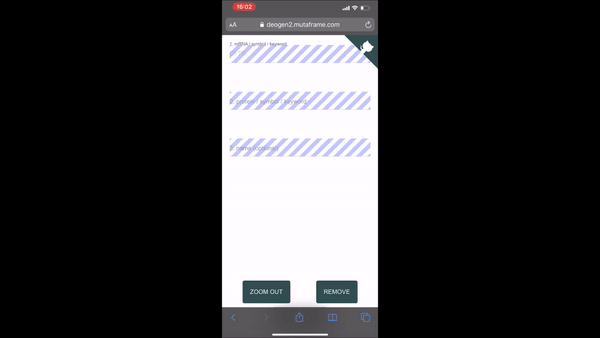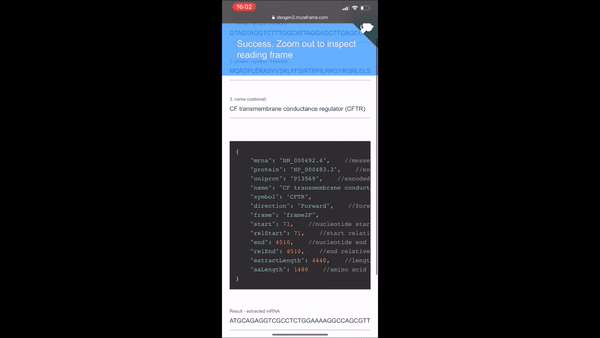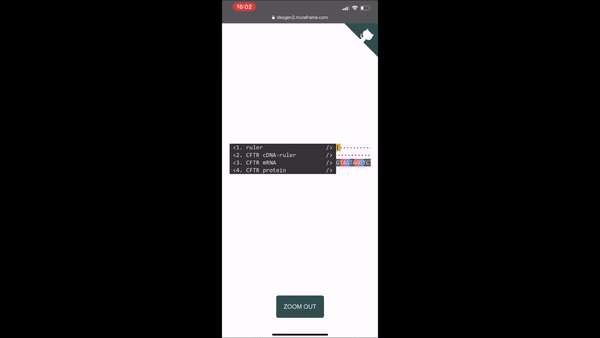
ORF (Open Reading Frame) is a sequence finder that relies on "pseudo" and "seqfetch" services. If you want to use the automated retrieved leave the mrna or protein section empty and type your keyword into the other.
Alternatively you can copy paste mrna and protein sequences on their respective lines and click "ANALYZE" to check if the given protein sequence is found within one of the six possible frames of the given mrna.
You can add as many sequences as you like, they will be justified to the left for comparison:
| field | desc | default | required |
|---|---|---|---|
| uniprot_accession | uniprot accession | NA | required |
| interval | interval | 1,sequence.length |
optional |
| fields | return fields | NA | required |
Array. Empty object if bad request is received
GET https://deogen2.mutaframe.com/tools/scorefetch?uniprot_accession=P19838&fields=pr,ci&interval=1,11-20,30-40
[{"name":"M1A","pr":"-0.806","ci":"0.1102190986535408"},{"name":"M1C","pr":"-1.169","ci":"0.1102190986535408"},{"name":"M1D","pr":"-1.563","ci":"0.1102190986535408"},{"name":"M1E","pr":"-1.274","ci":"0.1102190986535408"},{"name":"M1F","pr":"-0.995",....
You can track site and API status at https://status.mutaframe.com/
- Describe the issue in a few sentences
- If applicable, write down the UniprotID, if the bug appears in a specific protein
- Provide a screenshot of your console, by right clicking on an empty area and selecting "Inspect".
- Specify if the bug only appears on a certain browser or reproducible accross the board
- Describe the feature you'd like it to be added in a few sentences.
- Provide a link or a simple drawing which depicts feature if possible
- If you have suggestions for performance increase, take a screen shot of the area where the affected element is located
- If you have performance issues, describe the issue in a few sentences. If possible provide your rig's specs.
- If applicable, specifiy if the performance issue is present across all browsers, or confined to a specific one.
You can support Mutaframe by:
- following at
- creating an account at MutaFrame
- helping us create better software by submitting bug reports or feature requests