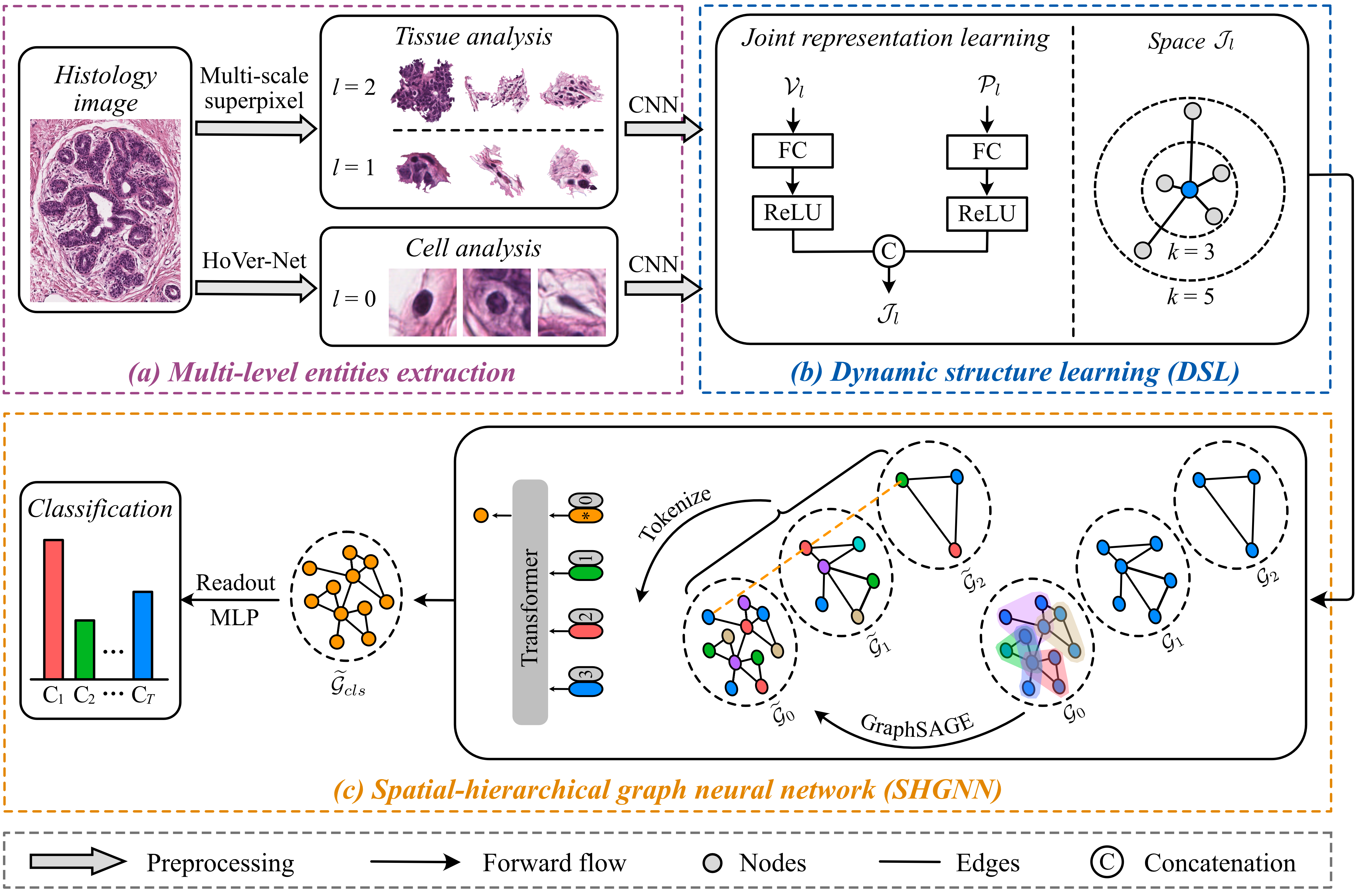
Spatial-hierarchical Graph Neural Network with Dynamic Structure Learning for Histological Image Classification
Installation
Clone the repo:
git clone https://github.com/HeLongHuang/SHGNN && cd SHGNNCreate a conda environment and activate it:
conda create -n env python=3.8
conda activate env
pip install -r requirements.txtData Preprocess
gendata.ipynb shows how to transform a histological image into the hierarchical graphs. After the data processing is completed, put all hierarchical graphs into a folder. The form is as follows:
PYG_Data
└── Dataset
├── pyg_data_1.pkl
├── pyg_data_2.pkl
:
└── pyg_data_n.pklYou also can download the processed graph data here. The access code is: dei3
Training
First, setting the data splits and hyperparameters in the file train.py. Then, experiments can be run using the following command-line:
cd train
python train_<experiments>.pyThe trained model will be saved in the folder SavedModels.
Saved models
We provide a 3-fold checkpoint for each dataset, which performing as:
| Dataset | Macro AUC |
|---|---|
| CRCS | 96.15 |
| BRACS | 95.06 |
Inference
Using the following command-line for model inference and result statistics:
cd inference
python inference_<experiments>.pyMore Info
-
Our implementation refers the following publicly available codes.
- Pytorch Geometric--Fey M, Lenssen J E. Fast graph representation learning with PyTorch Geometric[J]. arXiv preprint arXiv:1903.02428, 2019.
- Histocartography--Jaume G, Pati P, Anklin V, et al. HistoCartography: A toolkit for graph analytics in digital pathology[C]//MICCAI Workshop on Computational Pathology. PMLR, 2021: 117-128.
- Hact-net--Pati P, Jaume G, Foncubierta-Rodríguez A, et al. Hierarchical graph representations in digital pathology[J]. Medical image analysis, 2022, 75: 102264.
- ViT Pytorch--Dosovitskiy A, Beyer L, Kolesnikov A, et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale[C]//International Conference on Learning Representations. 2020.
-
Our in-house dataset (CRCS) cannot be made public due to the data use agreement and the protection of patients' privacy.
-
The BRACS dataset is available at:
- BRACS--Brancati N, Anniciello A M, Pati P, et al. Bracs: A dataset for breast carcinoma subtyping in h&e histology images[J]. arXiv preprint arXiv:2111.04740, 2021.
-
Results on BRACS with official data split:
-
Under the same experimental setup, we further validated our model on BRACS with official data split. The pretrained checkpoints of three random repeated experiments are provided, which performing as:
Metrics Macro AUC Weighted F1 Average 89.65 62.77
-
Citation
- If you found our work useful in your research, please consider citing our work at:
@InProceedings{10.1007/978-3-031-16434-7_18,
author="Hou, Wentai
and Huang, Helong
and Peng, Qiong
and Yu, Rongshan
and Yu, Lequan
and Wang, Liansheng",
title="Spatial-Hierarchical Graph Neural Network with Dynamic Structure Learning for Histological Image Classification",
booktitle="Medical Image Computing and Computer Assisted Intervention -- MICCAI 2022",
year="2022",
publisher="Springer Nature Switzerland",
address="Cham",
pages="181--191",
}