author: Xiong Wang
email: wangxiong@tjh.tjmu.edu.cn
affiliation: Department of Laboratory Medicine, Tongji Hospital, Tongji Medical College, HUST
Pan-cancer analysis aimed to examine the commonalities and heterogeneity among the genomic and cellular alterations across diverse types of tumors. Pan-cancer analysis of gene expression, tumor mutational burden (TMB), microsatellite instability (MSI), and tumor immune microenvironment (TIME) became available based on the exome, transcriptome, and DNA methylome data from TCGA. Some online tools provided user-friendly analysis of gene and protein expression, mutation, methylation, and survival for TCGA data, such as GEPIA 2 (http://gepia2.cancer-pku.cn/#index), cBioPortal (http://www.cbioportal.org/), UALCAN (https://ualcan.path.uab.edu/index.html), and MethSurv (https://biit.cs.ut.ee/methsurv/). However, these online tools were either uni-functional or not able to perform analysis of user-defined functions. Therefore, TCGA pan-cancer multi-omics data were integrated and included in this package, including gene expression TPM (transcripts per million) matrix, TMB, MSI, immune cell ratio, immune score, promoter methylation, and clinical information. A number of functions were generated to perform pan-cancer paired/unpaired differential gene expression analysis, pan-cancer correlation analysis between gene expression and TMB, MSI, immune cell ratio, immune score,immune stimulator,immune inhibitor, and promoter methylation. Methods for visualization were provided, including paired/unpaired boxplot, survival plot, ROC curve, heatmap, scatter, radar chart, and forest plot,in order to easily perform integrative pan-cancer multi-omics analysis. Finally, these built-in data could be extracted and analyzed with user-defined functions, making the pan-cancer analysis much more convenient.
To install this package, download TCGAplot R package at https://github.com/tjhwangxiong/TCGAplot/releases/download/v4.0.0/TCGAplot_4.0.0.zip and install locally.
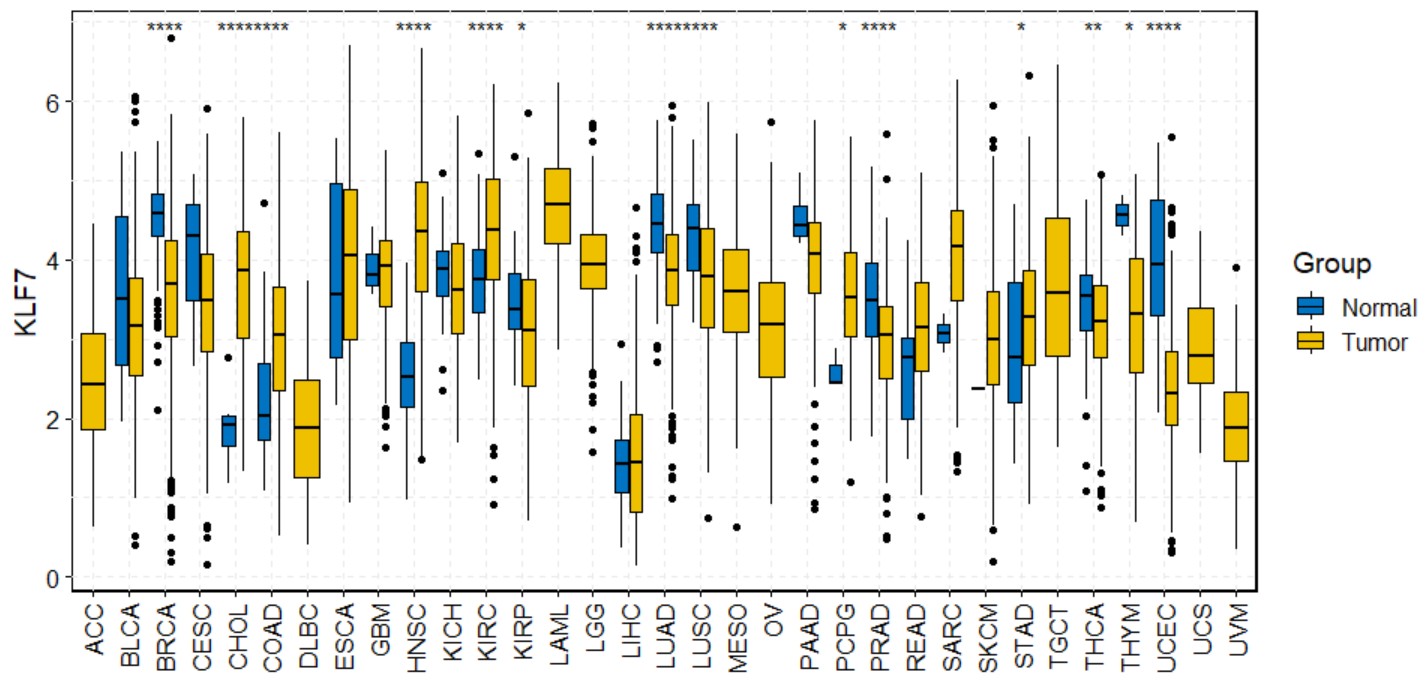
Create a pan-cancer box plot for a single gene with symbols indicating statistical significance.
pan_boxplot(gene,palette="jco",legend="right")gene
gene name likes "KLF7".
palette
the color palette to be used for filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco", "ucscgb", "uchicago", "simpsons" and "rickandmorty".
legend
legend position. Allowed values include "top","bottom","left","right" and "none".
Example
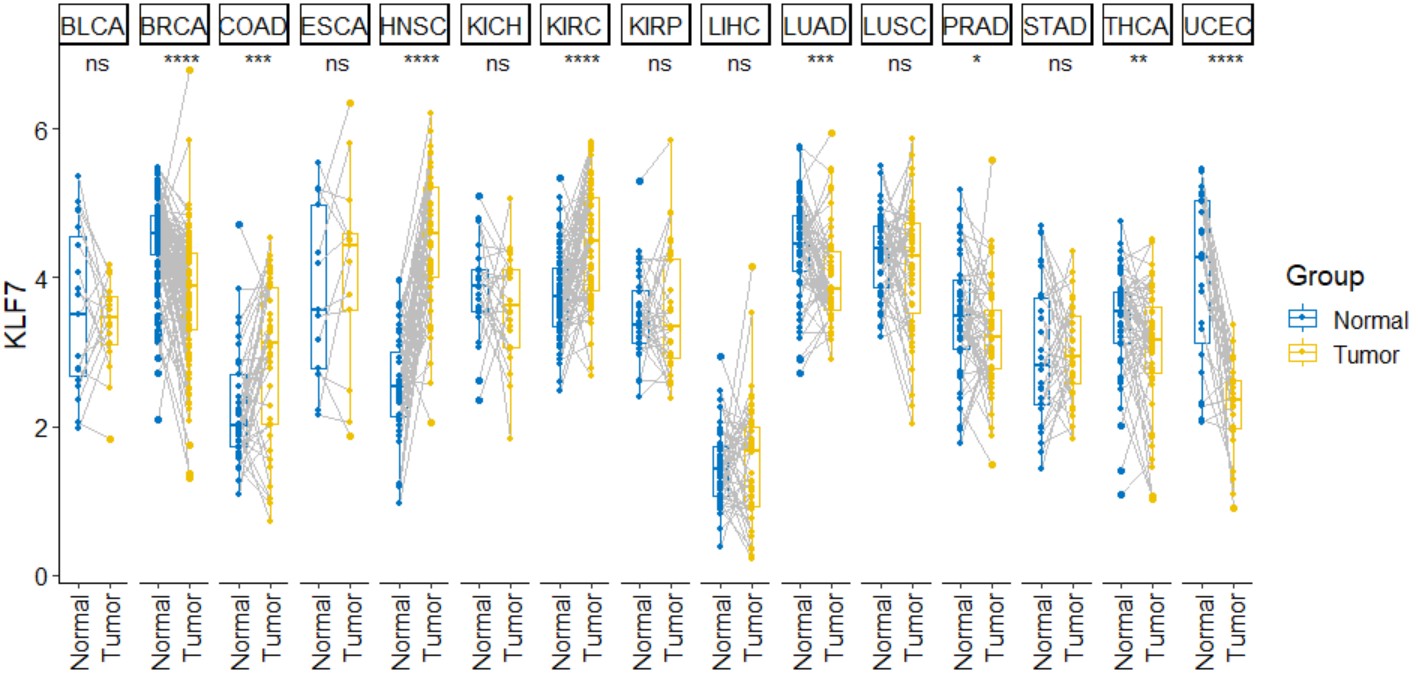
pan_boxplot("KLF7")Create a pan-cancer paired box plot for a single gene with symbols indicating statistical significance.
pan_paired_boxplot(gene,palette="jco",legend="right")gene
gene name likes "KLF7".
palette
the color palette to be used for filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco", "ucscgb", "uchicago", "simpsons" and "rickandmorty".
legend
legend position. Allowed values include "top","bottom","left","right" and "none".
Example
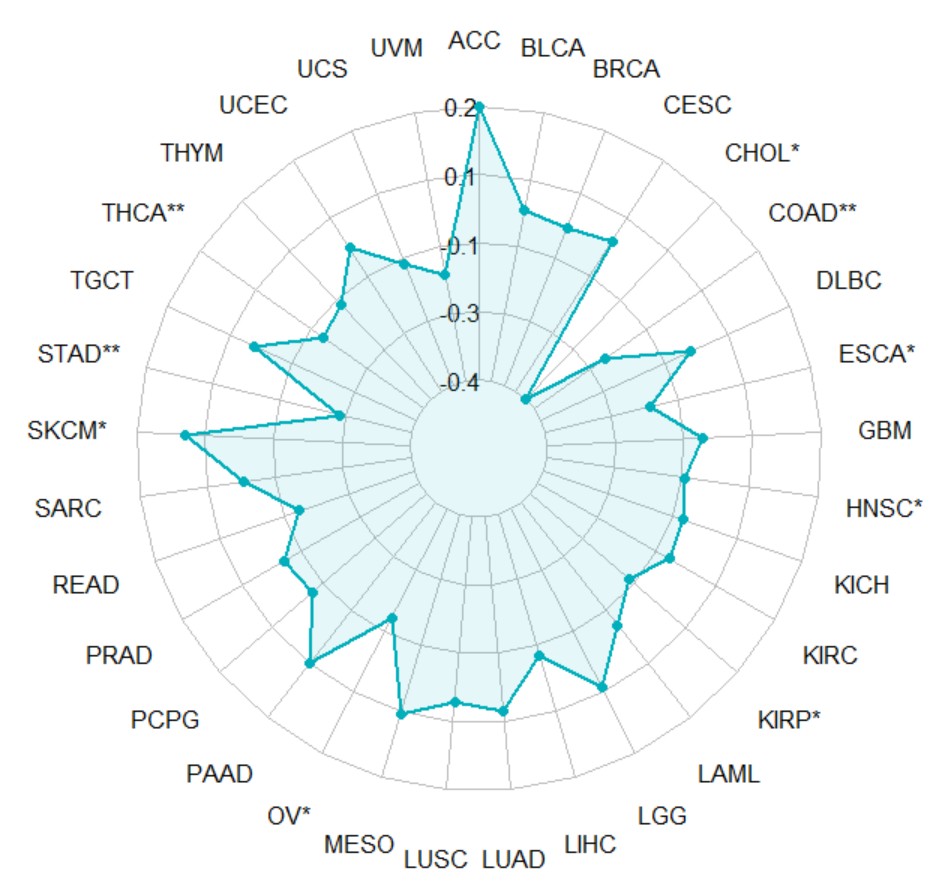
pan_paired_boxplot("KLF7")Create a pan-cancer radar chart for gene expression and TMB correlation.
gene_TMB_radar(gene,method = "pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
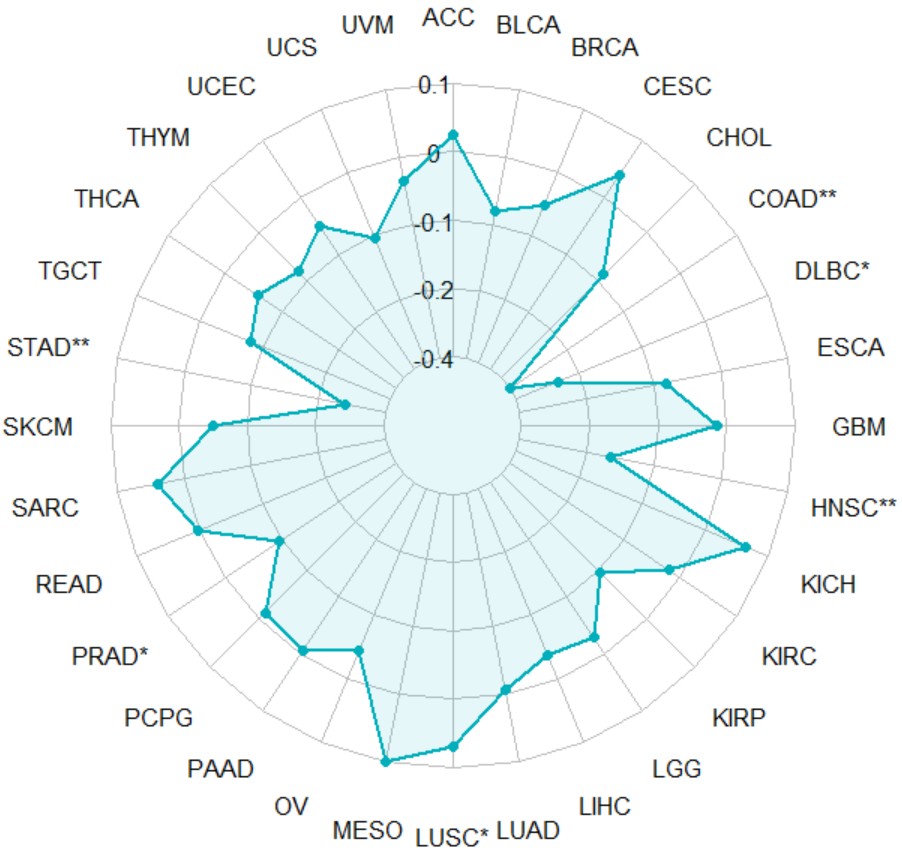
gene_TMB_radar("KLF7")Create a pan-cancer radar chart for gene expression and MSI correlation.
gene_MSI_radar(gene,method = "pearson")gene gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
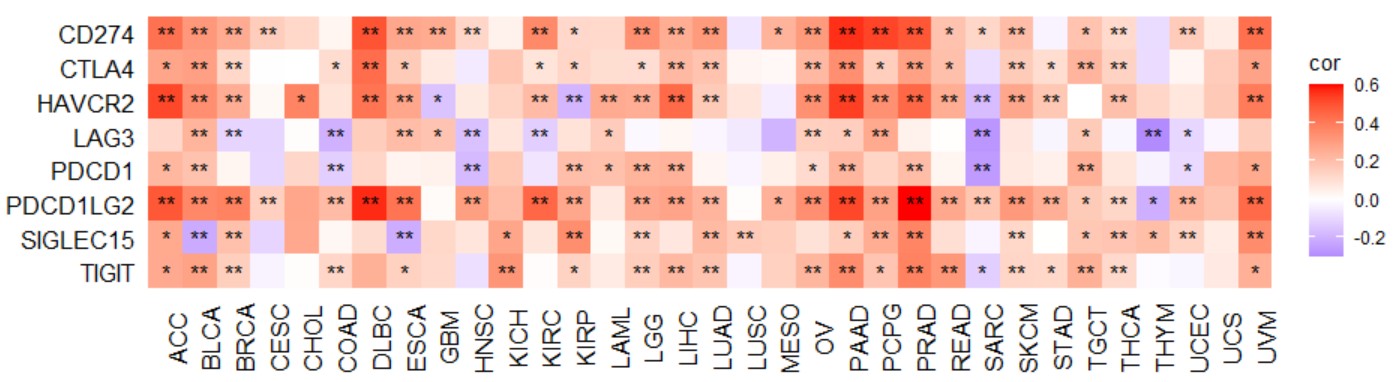
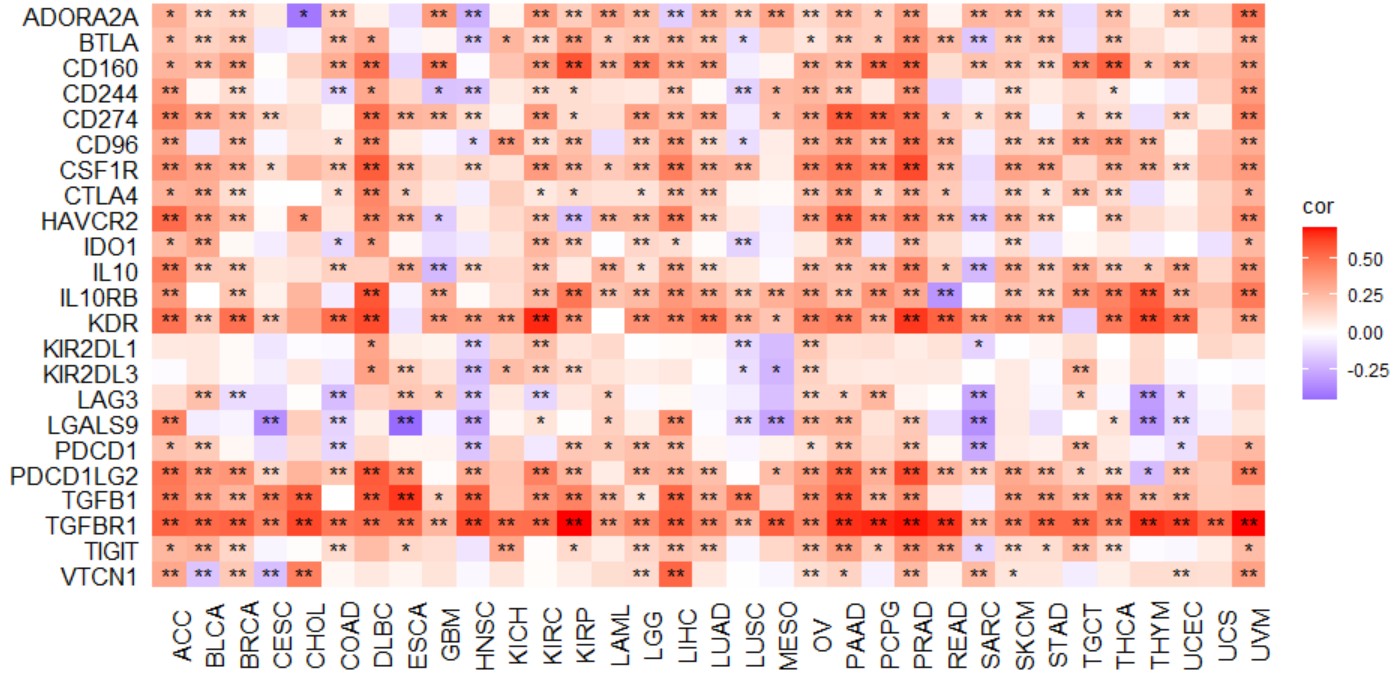
gene_MSI_radar("KLF7")Create a pan-cancer heatmap with symbols indicating statistical significance to reveal the correlation between the expression of a single gene and ICGs (immune checkpoint genes).
ICGs geneset included "CD274","CTLA4","HAVCR2","LAG3","PDCD1","PDCD1LG2","SIGLEC15",and "TIGIT".
gene_checkpoint_heatmap(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
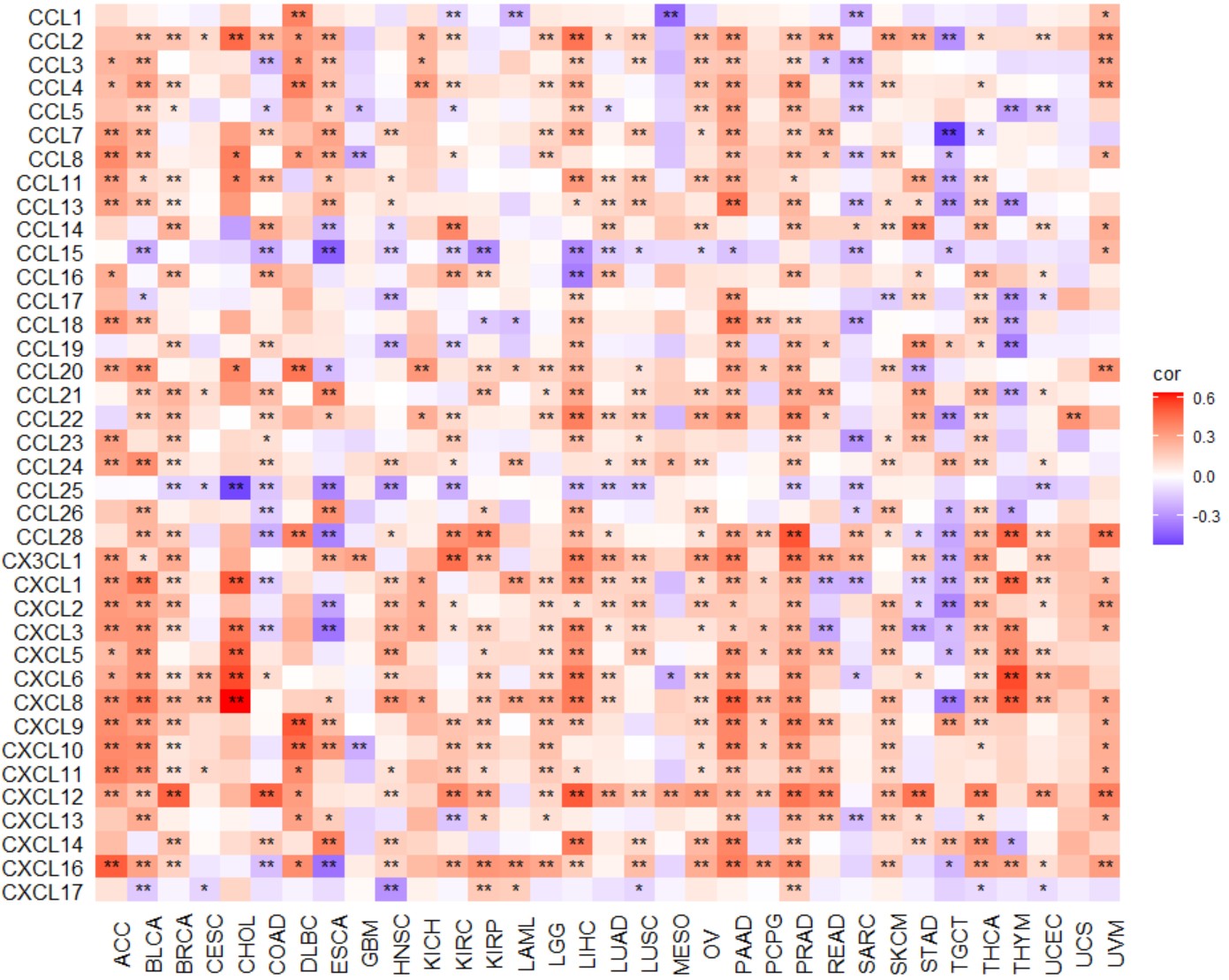
gene_checkpoint_heatmap("KLF7")Create a pan-cancer heatmap with symbols indicating statistical significance to reveal the correlation between the expression of a single gene and chemokine.
Chemokine geneset included "CCL1","CCL2","CCL3","CCL4","CCL5","CCL7","CCL8","CCL11","CCL13","CCL14","CCL15","CCL16","CCL17","CCL18","CCL19","CCL20","CCL21","CCL22","CCL23","CCL24","CCL25","CCL26","CCL28","CX3CL1","CXCL1","CXCL2","CXCL3","CXCL5","CXCL6","CXCL8","CXCL9","CXCL10","CXCL11","CXCL12","CXCL13","CXCL14","CXCL16", and "CXCL17".
gene_chemokine_heatmap(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
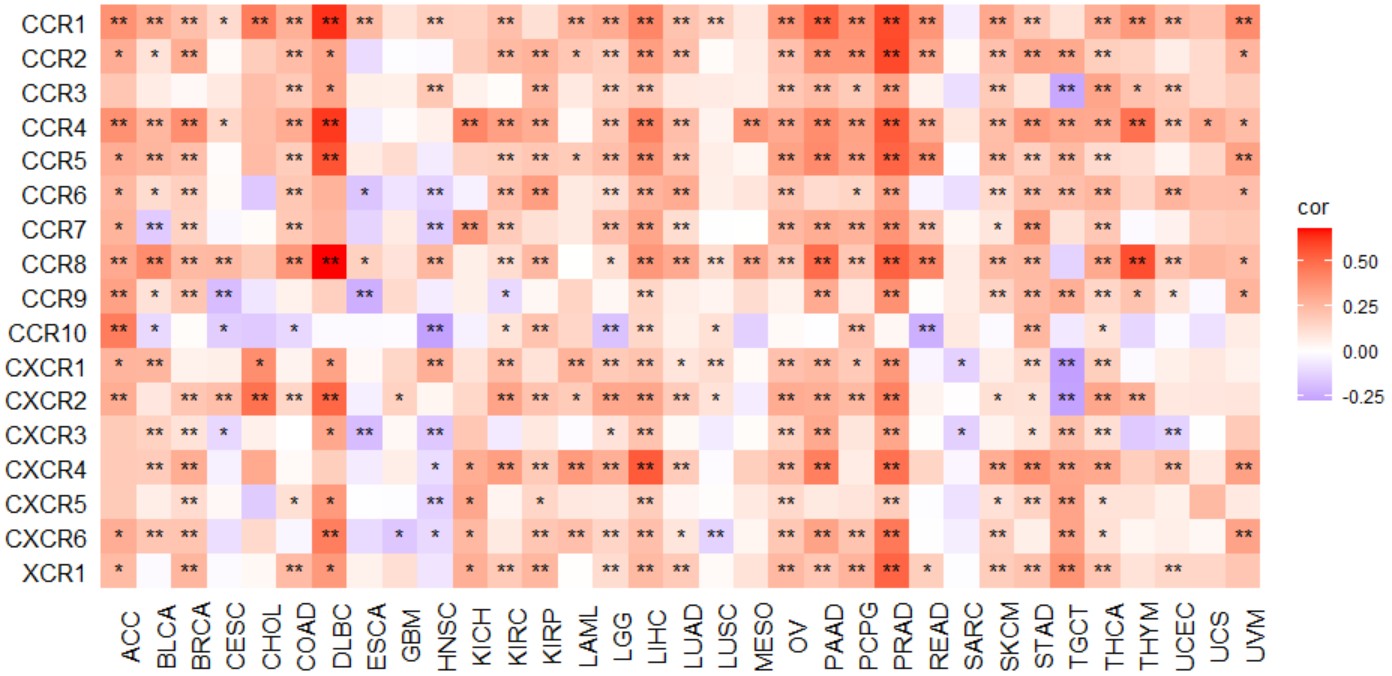
gene_chemokine_heatmap("KLF7")Create a pan-cancer heatmap with symbols indicating statistical significance to reveal the correlation between the expression of a single gene and chemokine receptors.
Chemokine receptor geneset included "CCR1","CCR2","CCR3","CCR4","CCR5","CCR6","CCR7","CCR8","CCR9","CCR10", "CXCR1","CXCR2","CXCR3","CXCR4","CXCR5","CXCR6","XCR1", and "CX3R1".
gene_receptor_heatmap(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
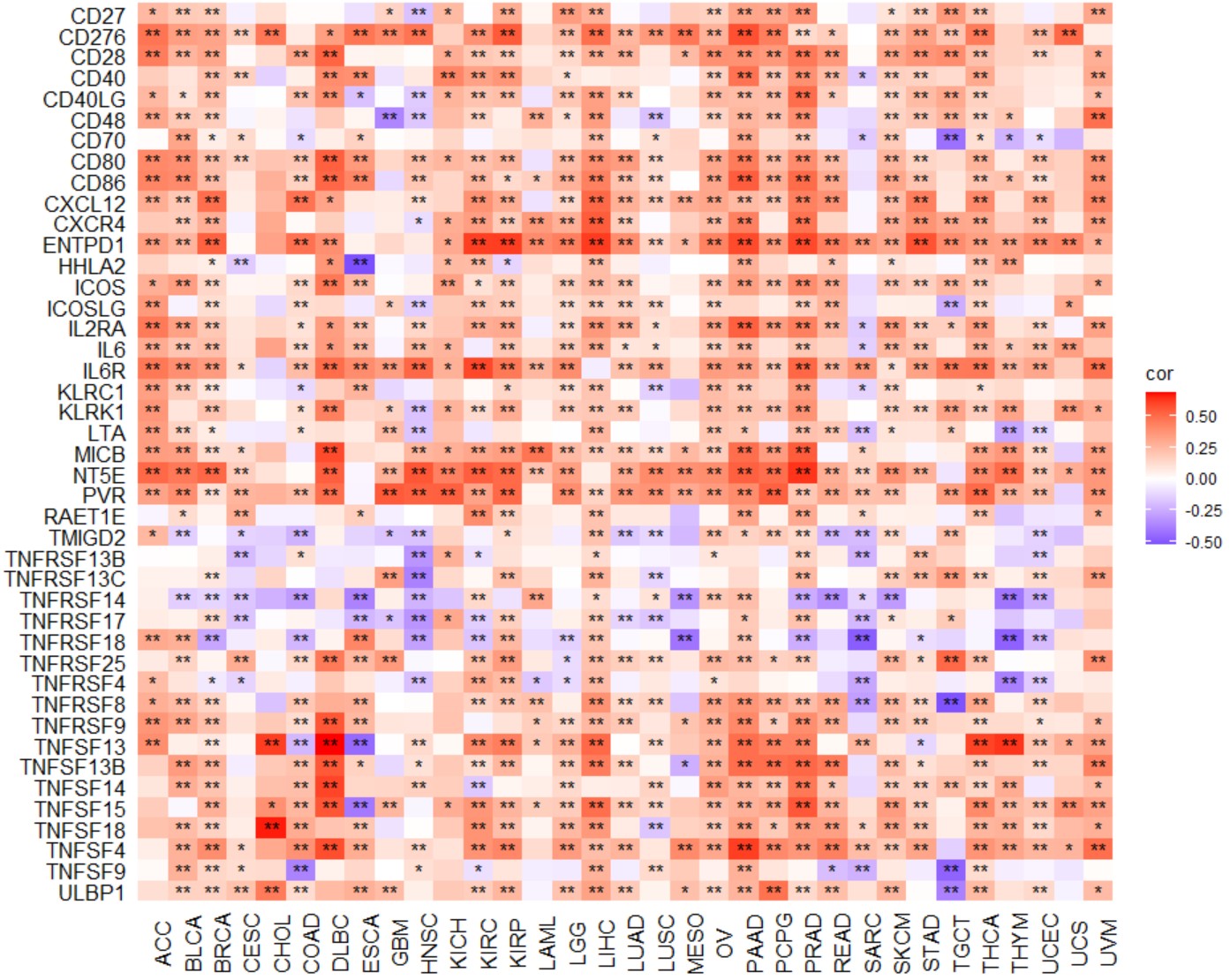
gene_receptor_heatmap("KLF7")Create a pan-cancer heatmap with symbols indicating statistical significance to reveal the correlation between the expression of a single gene and immune stimulators.
Immune stimulator geneset included "CD27","CD276","CD28","CD40","CD40LG","CD48","CD70","CD80","CD86","CXCL12","CXCR4","ENTPD1","HHLA2","ICOS","ICOSLG","IL2RA","IL6","IL6R","KLRC1","KLRK1","LTA","MICB","NT5E","PVR","RAET1E","TMIGD2","TNFRSF13B","TNFRSF13C","TNFRSF14","TNFRSF17","TNFRSF18","TNFRSF25","TNFRSF4","TNFRSF8","TNFRSF9","TNFSF13","TNFSF13B","TNFSF14","TNFSF15","TNFSF18","TNFSF4","TNFSF9", and "ULBP1".
gene_immustimulator_heatmap(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
gene_immustimulator_heatmap("KLF7")Create a pan-cancer heatmap with symbols indicating statistical significance to reveal the correlation between the expression of a single gene and immune inhibitors.
Immune inhibitor geneset included "ADORA2A","BTLA","CD160","CD244","CD274","CD96","CSF1R","CTLA4","HAVCR2","IDO1","IL10","IL10RB","KDR","KIR2DL1","KIR2DL3","LAG3","LGALS9","PDCD1","PDCD1LG2","TGFB1","TGFBR1","TIGIT", and "VTCN1".
gene_immuinhibitor_heatmap(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
gene_immuinhibitor_heatmap("KLF7")Create a pan-cancer heatmap with symbols indicating statistical significance to reveal the correlation between the expression of a single gene and immune cell ratio.
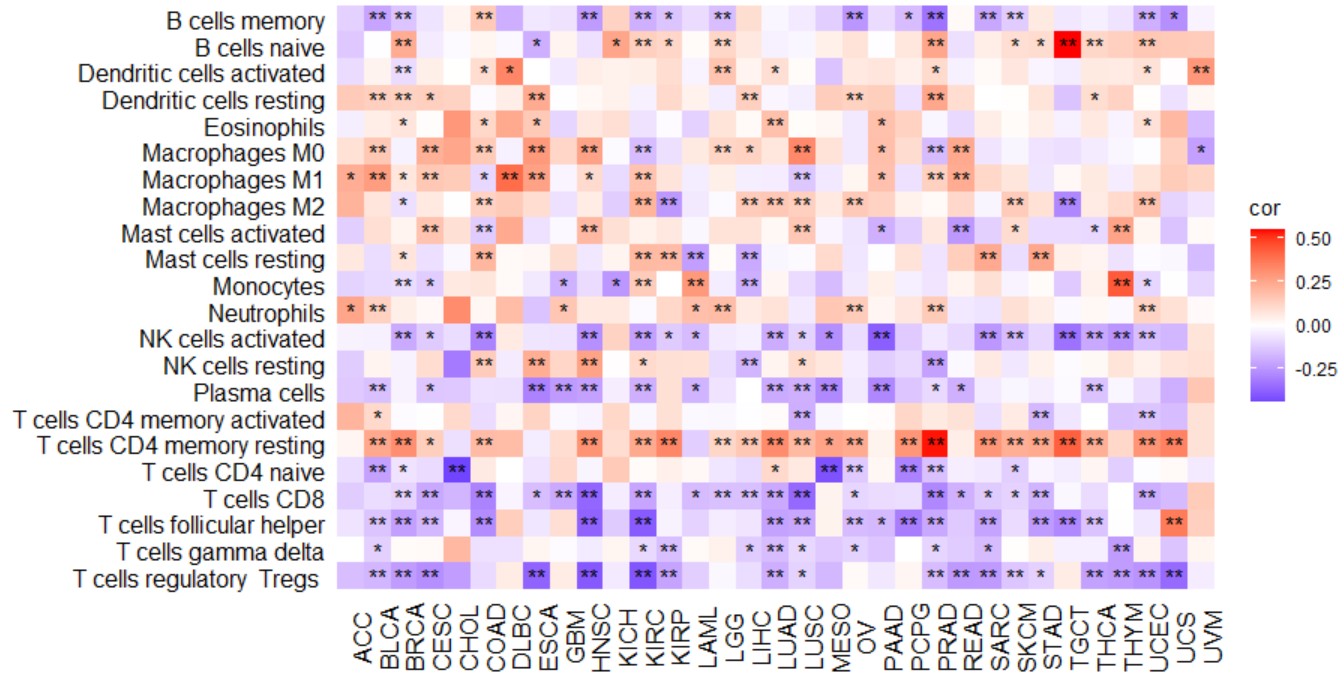
gene_immucell_heatmap(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
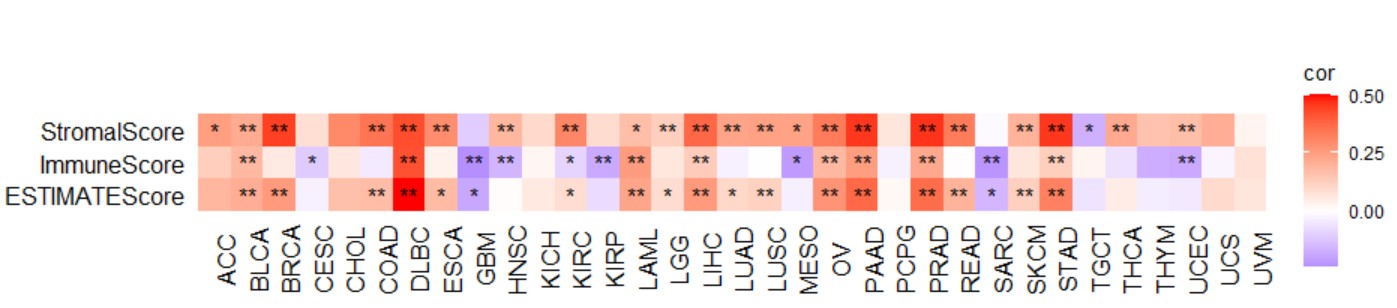
gene_immucell_heatmap("KLF7")Create a pan-cancer heatmap with symbols indicating statistical significance to reveal the correlation between the expression of a single gene and immune scores, including Stromal score, immune score, and ESTIMATE score.
gene_immunescore_heatmap(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
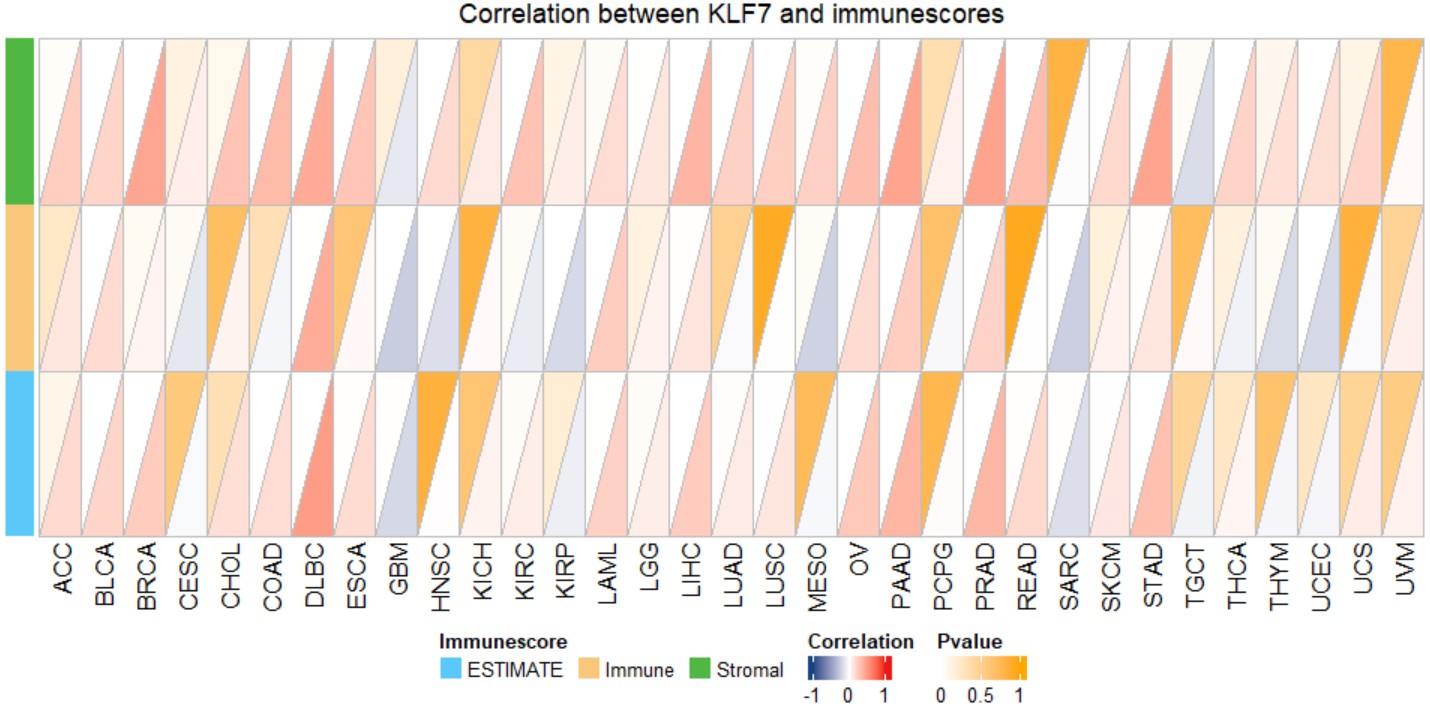
gene_immunescore_heatmap("KLF7")Create a pan-cancer triangle reveals the correlation between the expression of a single gene and immune scores, including Stromal score, immune score, and ESTIMATE score.
gene_immunescore_triangle(gene,method="pearson")gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
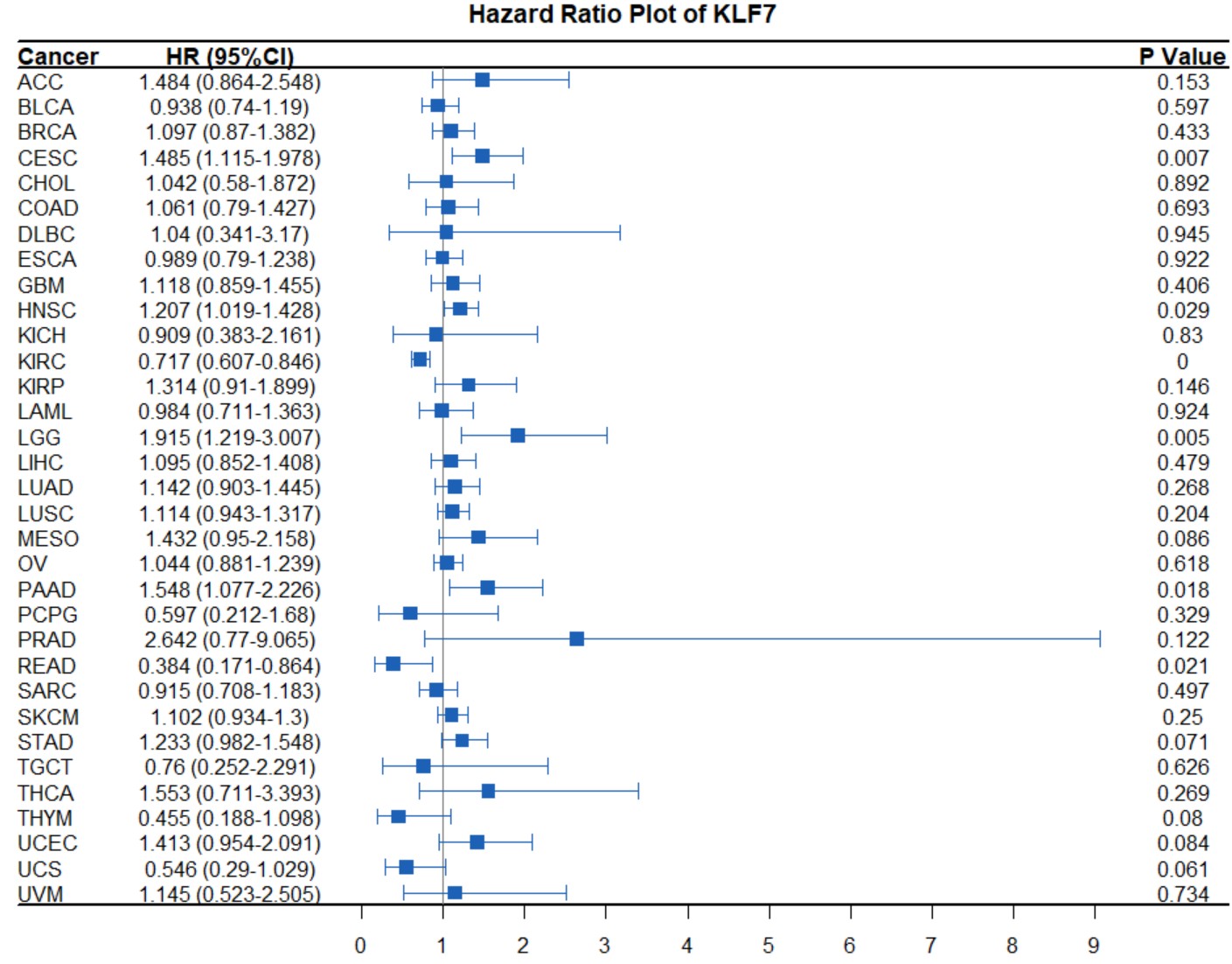
gene_immunescore_triangle("KLF7")Create a pan-cancer Cox regression forest plot for a specific gene.
pan_forest(gene)gene
gene name likes "KLF7".
method
method="pearson" is the default value. The alternatives to be passed to correlation are "spearman" and "kendall".
Example
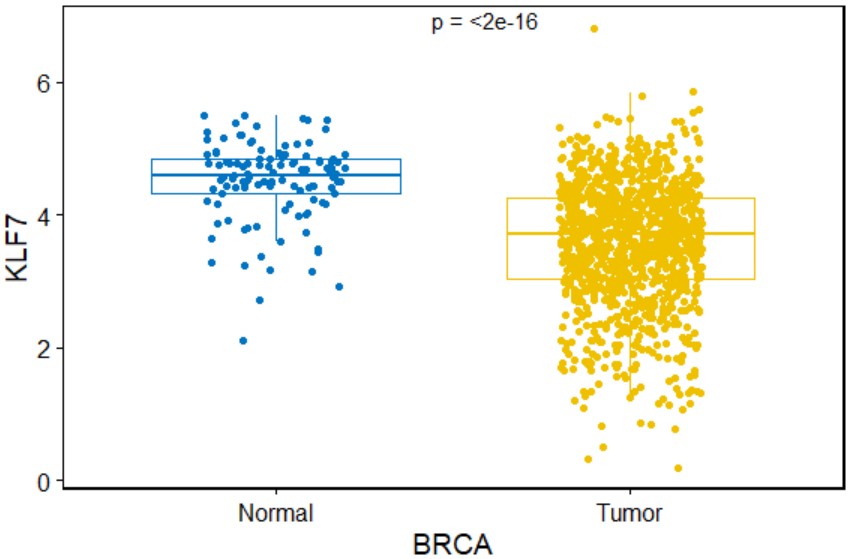
pan_forest("KLF7")Create a tumor-normal box plot for a single gene with symbols indicating statistical significance in a specific type of cancer.
tcga_boxplot(cancer,gene,add = "jitter",palette="jco",legend="none")cancer
cancer name likes "BRCA".
gene
gene name likes "KLF7".
add
character vector for adding another plot element likes "none", "dotplot", "jitter".
palette
the color palette to be used for coloring or filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco".
legend
legend position. Allowed values include "top","bottom","left","right" and "none".
Example
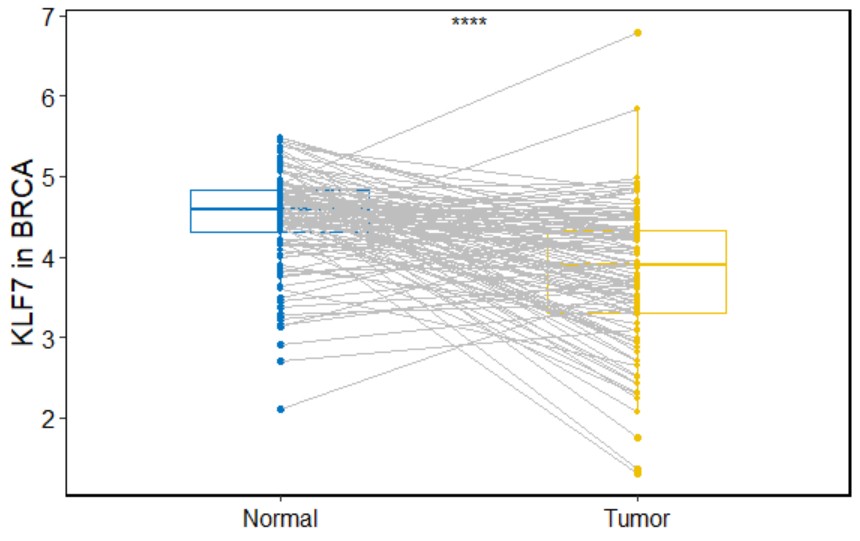
tcga_boxplot("BRCA","KLF7")Create a paired tumor-normal box plot for a single gene with symbols indicating statistical significance in a specific type of cancer.
Only cancers with more than 20 paired samples could be analyzed, including "BLCA","BRCA","COAD","ESCA","HNSC","KICH","KIRC","KIRP","LIHC","LUAD","LUSC","PRAD","STAD","THCA", and "UCEC".
paired_boxplot(cancer,gene,palette="jco",legend="none")cancer
cancer name likes "BRCA".
gene
gene name likes "KLF7".
palette
the color palette to be used for coloring or filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco".
legend
legend position. Allowed values include "top","bottom","left","right" and "none".
Example
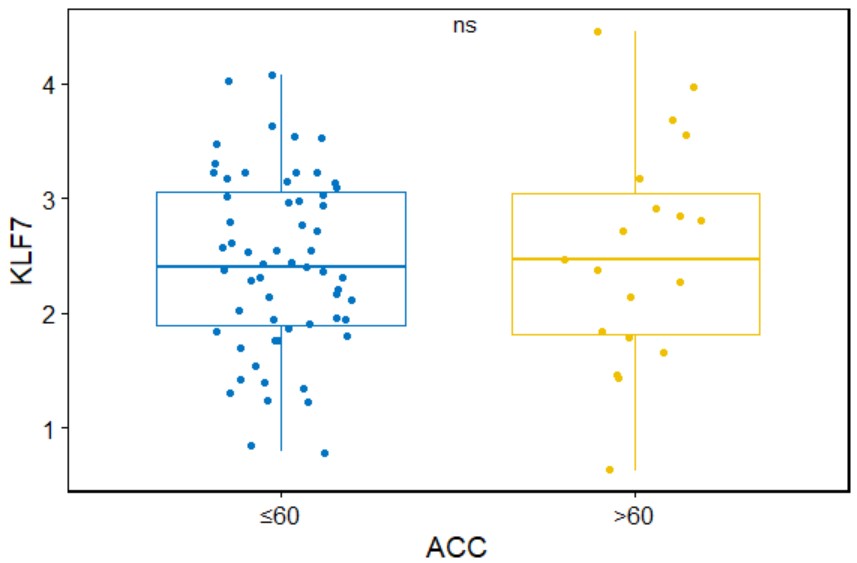
paired_boxplot("BRCA","KLF7")Create a box plot for a single gene with symbols indicating statistical significance grouped by age in a specific type of cancer.
gene_age(cancer,gene,age=60,add = "jitter",palette="jco",legend="none")cancer
cancer name likes "ACC".
gene
gene name likes "KLF7".
age
numeric number of age like 60.
add
character vector for adding another plot element likes "none", "dotplot", "jitter".
palette
the color palette to be used for coloring or filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco".
legend
legend position. Allowed values include "top","bottom","left","right" and "none".
Example
gene_age("ACC","KLF7")Create a box plot for a single gene with symbols indicating statistical significance grouped by gender in a specific type of cancer.
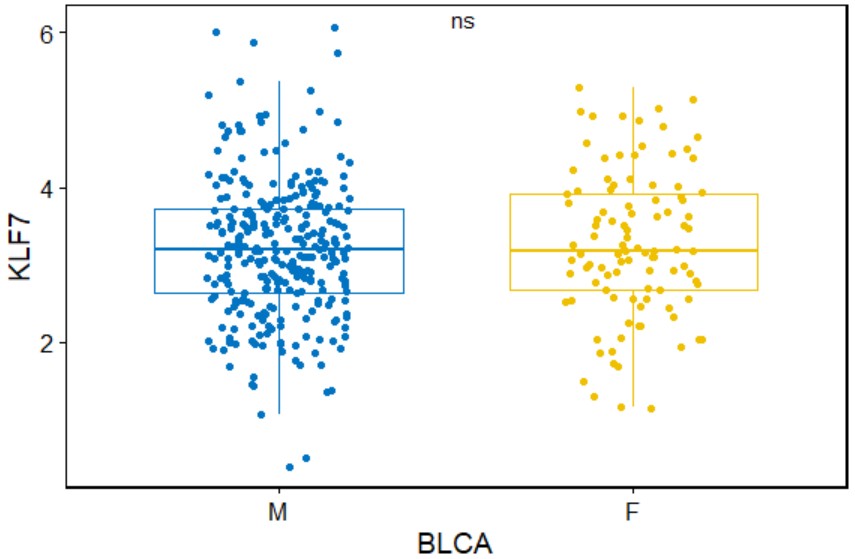
gene_gender(cancer,gene,add = "jitter",palette="jco",legend="none")cancer
cancer name likes "BLCA".
gene
gene name likes "KLF7".
add
character vector for adding another plot element likes "none", "dotplot", "jitter".
palette
the color palette to be used for coloring or filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco".
legend
legend position. Allowed values include "top","bottom","left","right" and "none".
Example
gene_gender("BLCA","KLF7")Create a box plot for a single gene with symbols indicating statistical significance grouped by stage in a specific type of cancer.
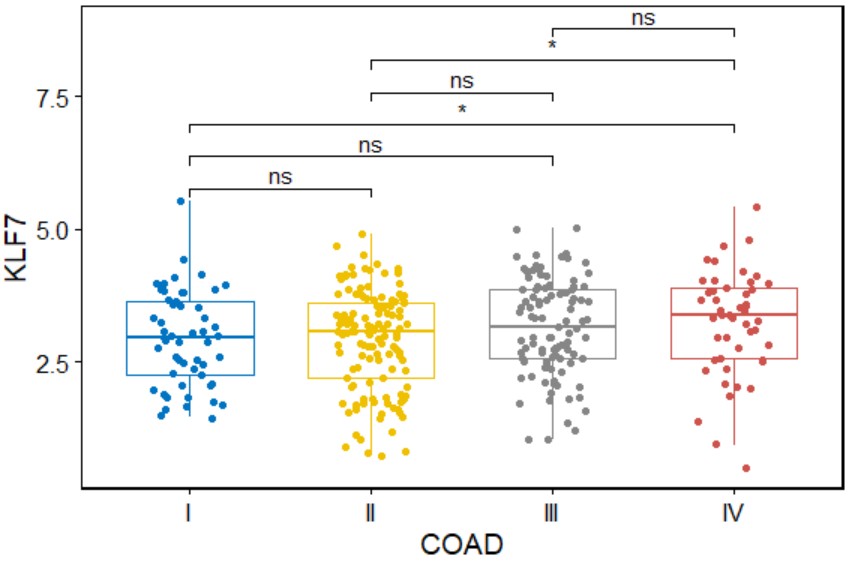
gene_gender(cancer,gene,add = "jitter",palette="jco",legend="none")cancer
cancer name likes "COAD".
gene
gene name likes "KLF7".
add
character vector for adding another plot element likes "none", "dotplot", "jitter".
palette
the color palette to be used for coloring or filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco".
legend
legend position. Allowed values include "top","bottom","left","right" and "none".
Example
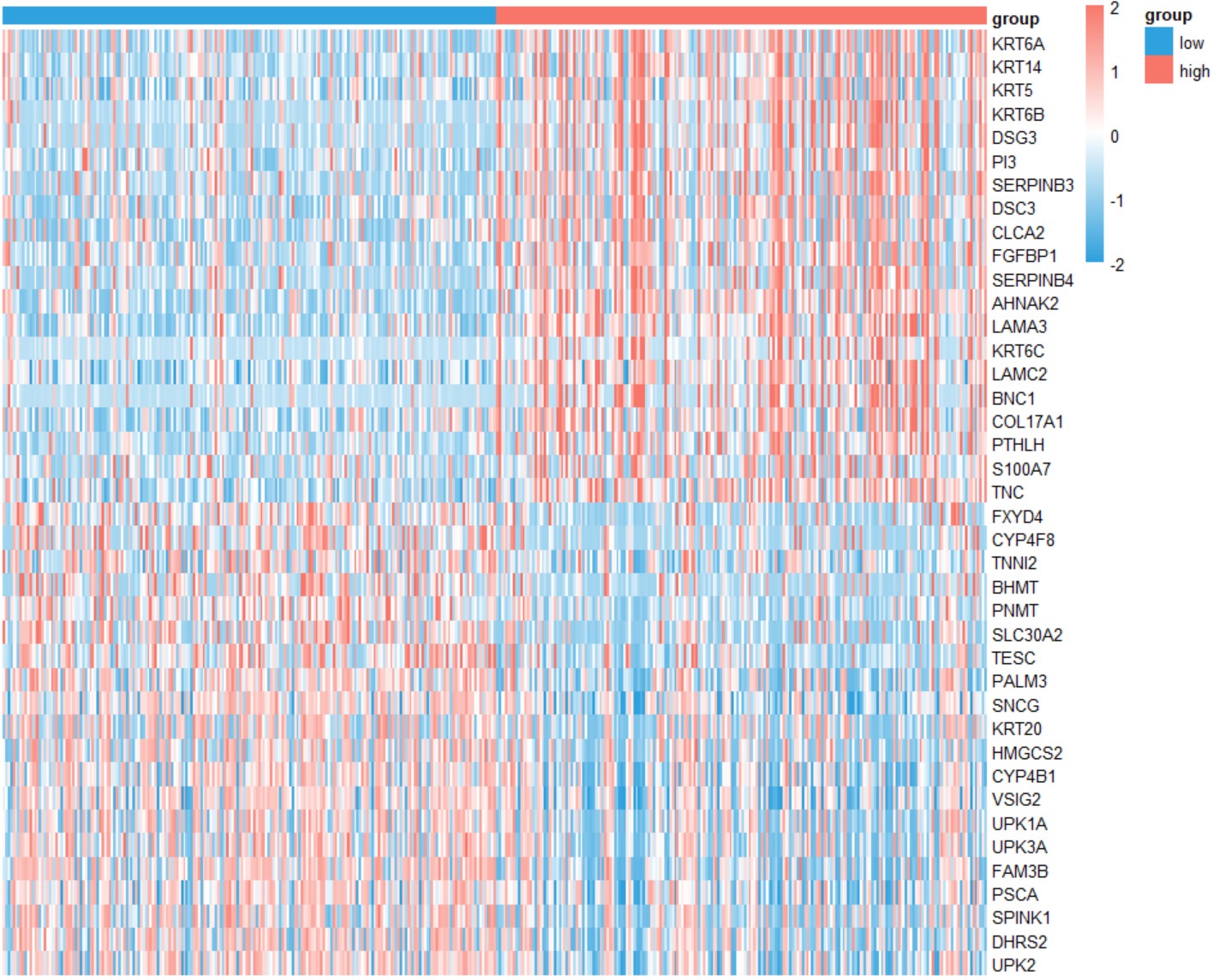
gene_stage("COAD","KLF7")Create a heatmap for differentially expressed genes grouped by the expression of a single gene in a specific type of cancer.
gene_deg_heatmap(cancer, gene,top_n=20)cancer
cancer name likes "BLCA".
gene
gene name likes "KLF7".
top_n
the number of top DEGS to be shown in the heatmap.
Example
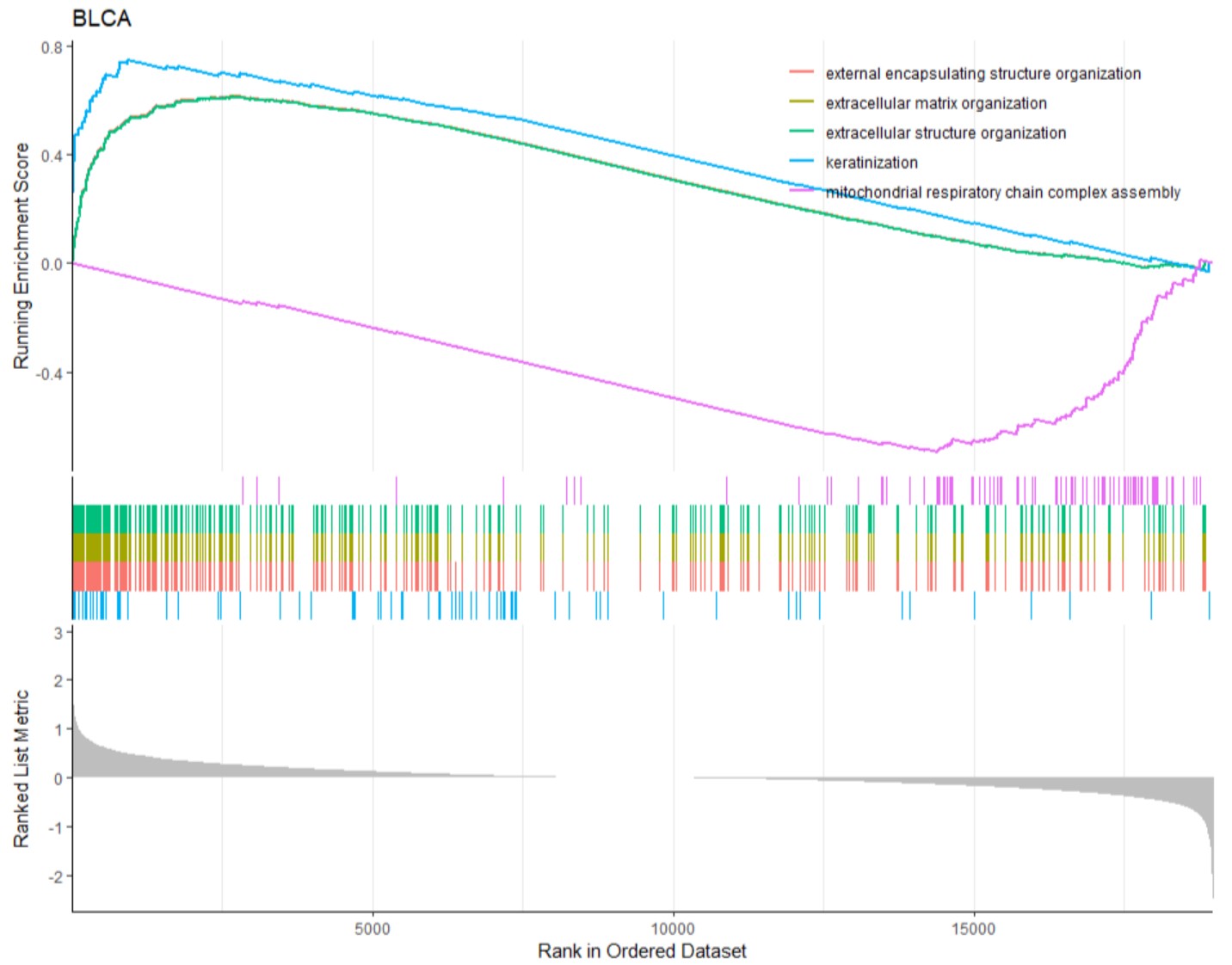
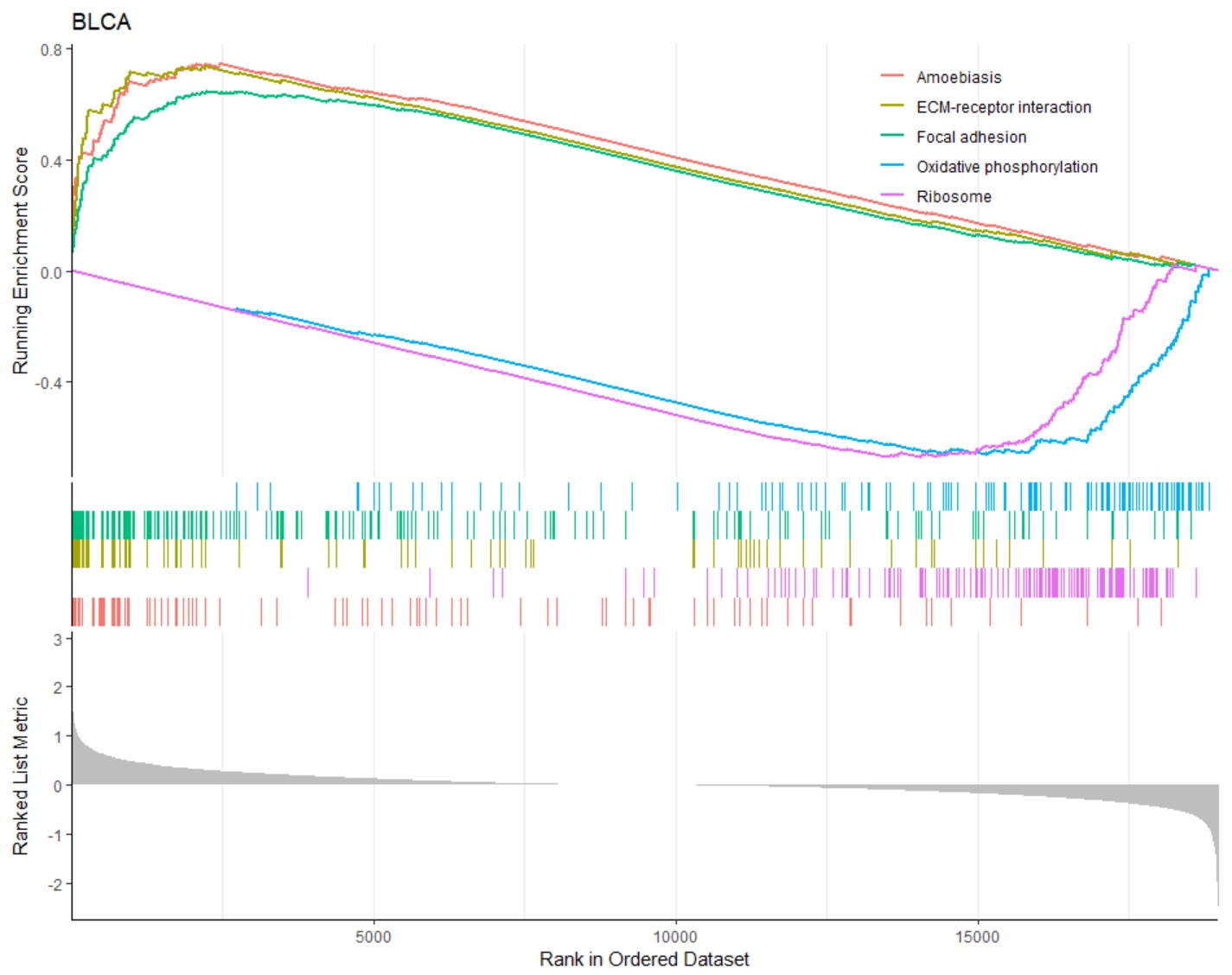
gene_deg_heatmap("BLCA","KLF7")GSEA-GO analysis of DEGs grouped by the expression of a single gene in a specific type of cancer, and the top 5 GO BP pathways were shown.
gene_gsea_go(cancer,gene,logFC_cutoff=2,pvalue_cutoff = 0.05)cancer
cancer name likes "BLCA".
gene
gene name likes "KLF7".
logFC_cutoff cutoff value of logFC, 2 was the default setting.
pvalue_cutoff
cutoff value of pvalue, 0.05 was the default setting.
Example
gene_gsea_go("BLCA","KLF7")GSEA-KEGG analysis of DEGs grouped by the expression of a single gene in a specific type of cancer, and the top 5 KEGG pathways were shown.
gene_gsea_kegg(cancer,gene,logFC_cutoff=2,pvalue_cutoff = 0.05)cancer
cancer name likes "BLCA".
gene
gene name likes "KLF7".
logFC_cutoff cutoff value of logFC, 2 was the default setting.
pvalue_cutoff
cutoff value of pvalue, 0.05 was the default setting.
Example
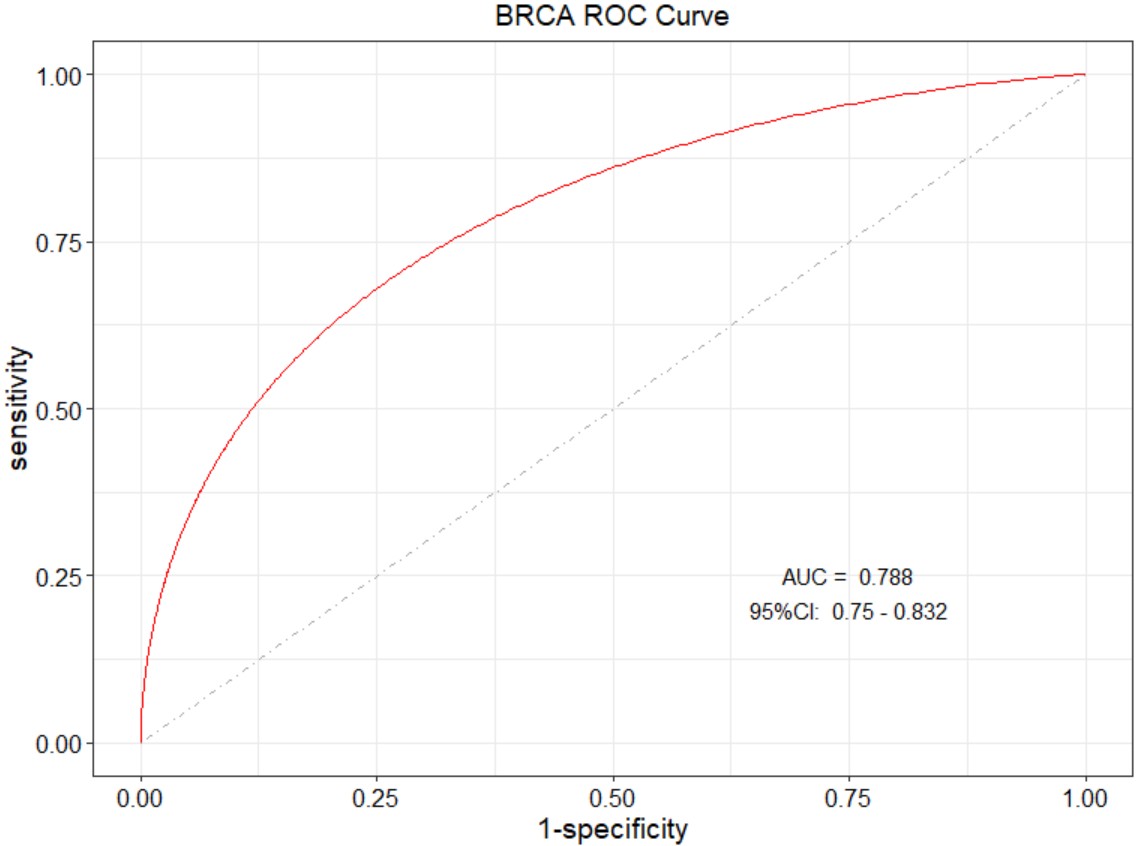
gene_gsea_kegg("BLCA","KLF7")Diagnostic ROC curve of a single gene in a specific type of cancer.
tcga_roc(cancer,gene)cancer
cancer name likes "BRCA".
gene
gene name likes "KLF7".
Example
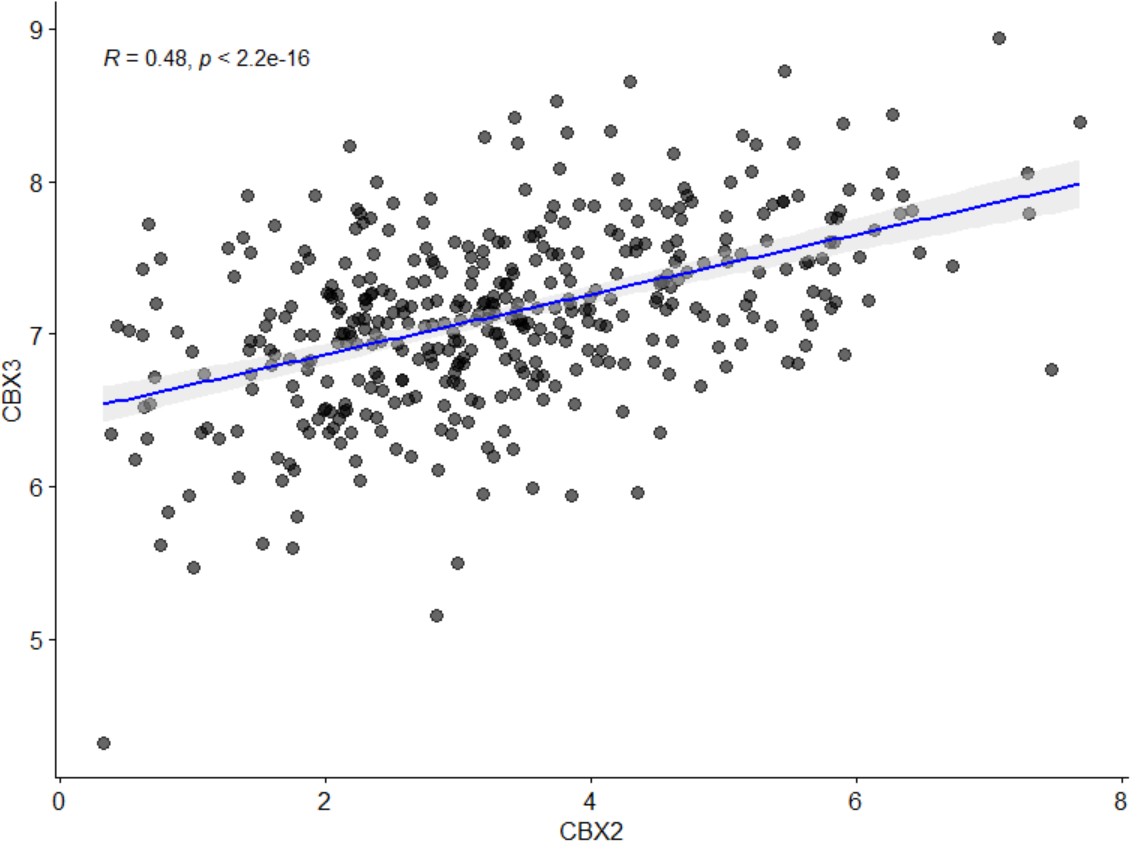
tcga_roc("BRCA","KLF7")Scatter plot of gene and gene correlation in a specific type cancer.
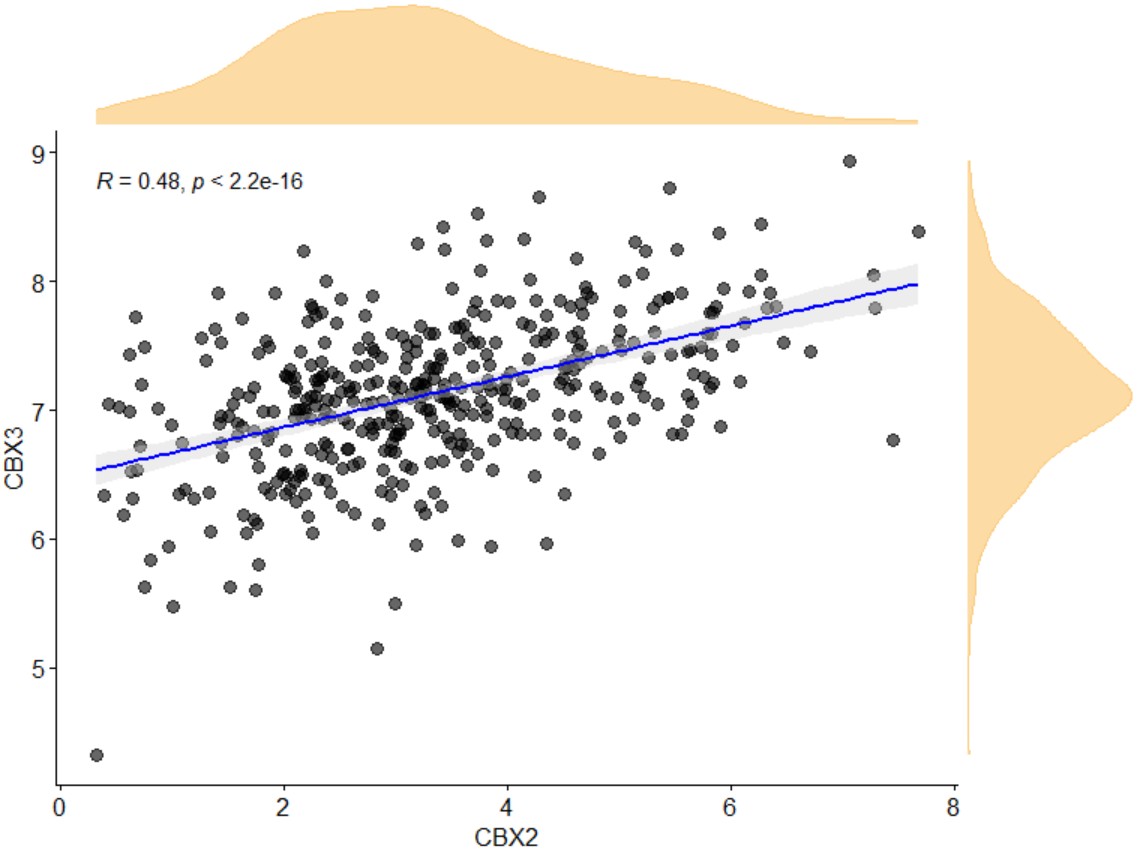
gene_gene_scatter(cancer,gene1,gene2,density="F")cancer
cancer name likes "BLCA".
gene1
name of gene1 likes "CBX2".
gene2
name of gene1 likes "CBX3".
density
whether density of gene expression was shown.
Example
gene_gene_scatter("BLCA","CBX2","CBX3")
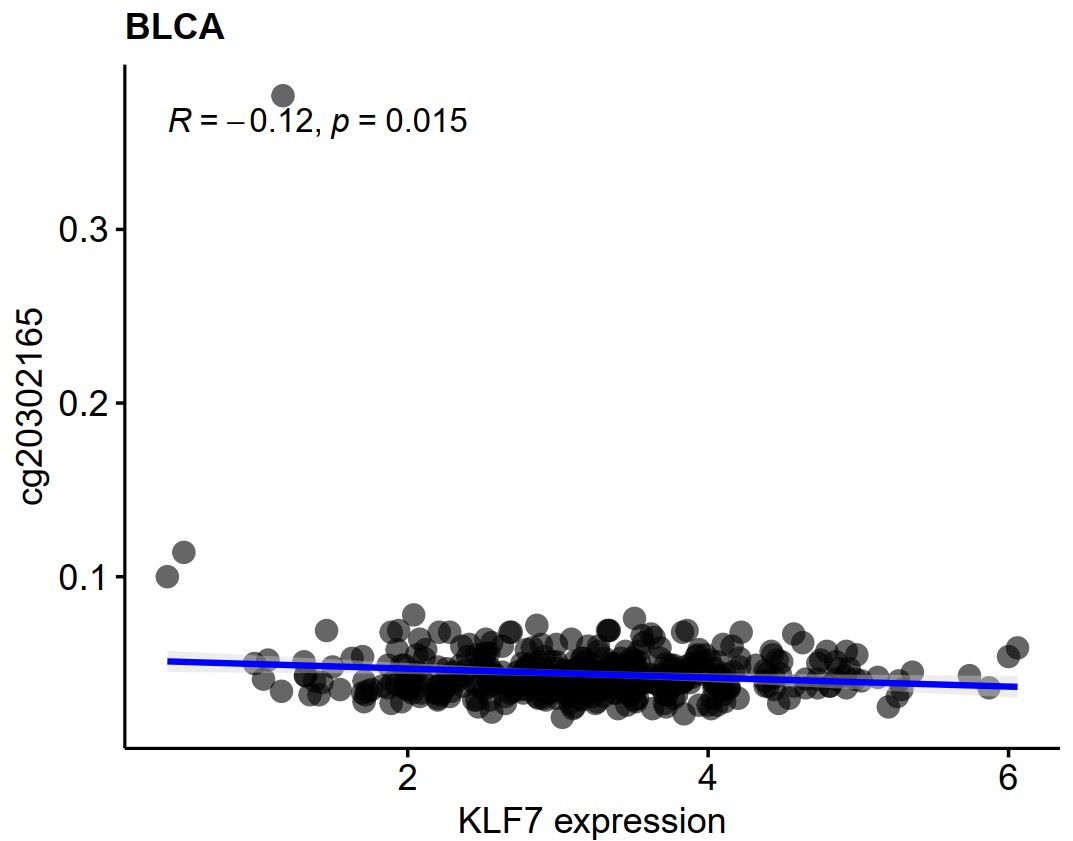
gene_gene_scatter("BLCA","CBX2","CBX3",density="T")Scatter plot of gene expression and gene promoter methylation correlation in a specific type of cancer. A pdf file named gene_methylation will be generated in the working directory.
gene_methylation_scatter(cancer,gene)cancer
cancer name likes "BLCA".
gene
gene name likes "KLF7".
Example
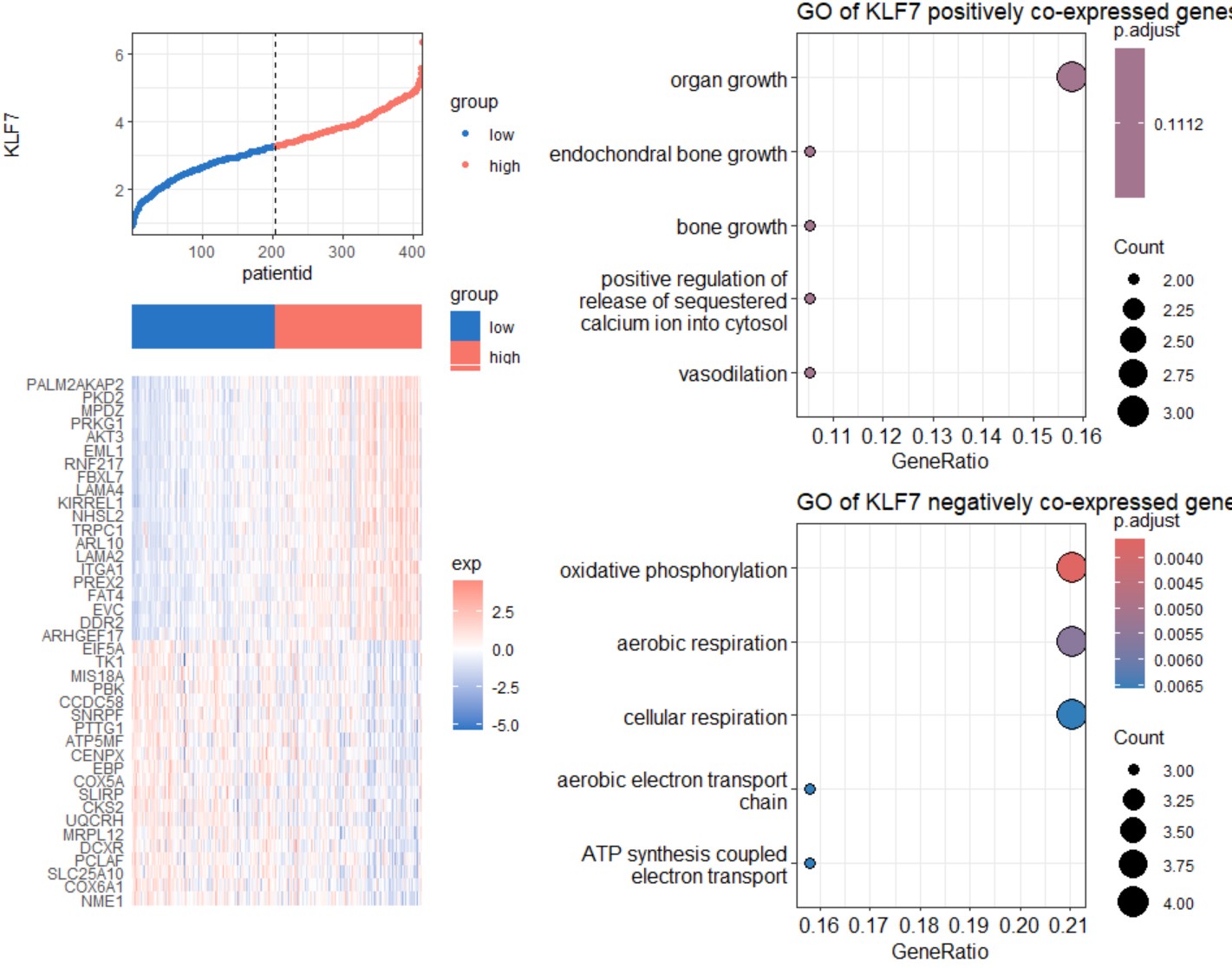
gene_methylation_scatter("BLCA","KLF7")Heatmap and Go enrichment of the positive and negative co-expressed genes of a single gene in a specific type of cancer.
gene_coexp_heatmap(cancer,gene,top_n=20, method="pearson")cancer
cancer name likes "STAD".
gene
gene name likes "KLF7".
top_n the number of co-expressed genes.
method method="pearson" is the default value. The alternatives to be passed to correlation were "spearman" and "kendall".
Example
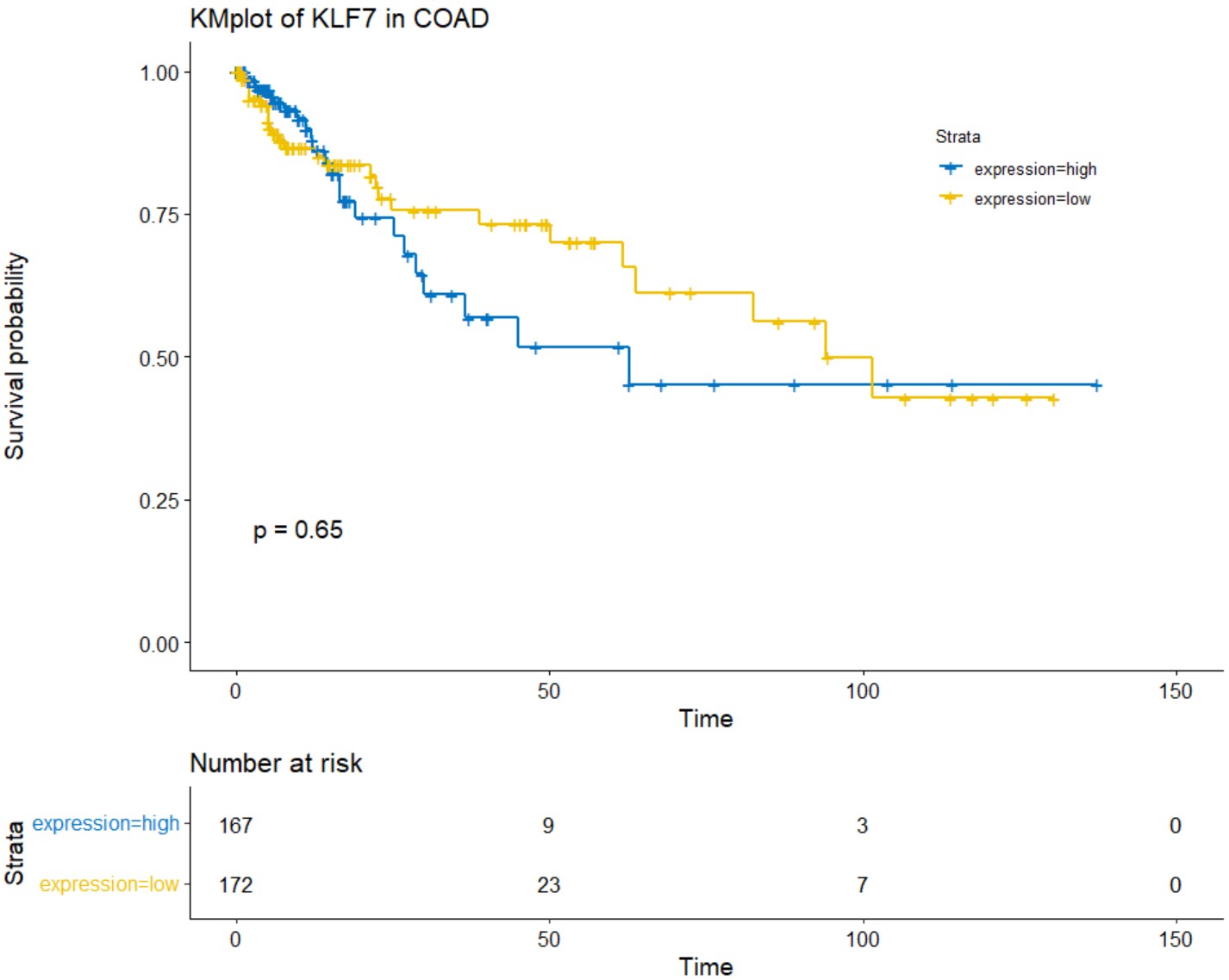
gene_coexp_heatmap("STAD","KLF7")K_M survival plot for a single gene in a specific type of cancer.
tcga_kmplot(cancer,gene,palette='jco')cancer
cancer name likes "COAD".
gene
gene name likes "KLF7".
palette the color palette to be used for coloring or filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco".
Example
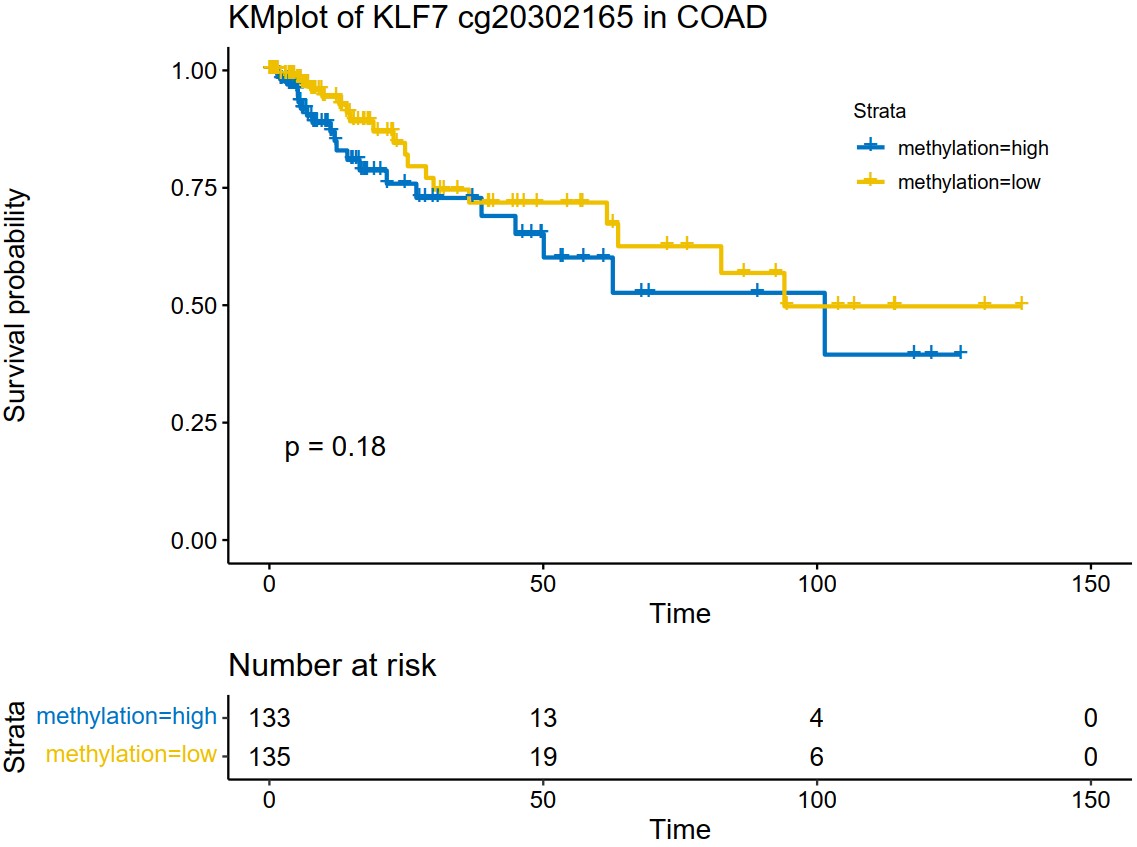
tcga_kmplot("COAD","KLF7")Describes the K_M survival plot based on the promoter methylation of a single gene in a specific type of cancer. A pdf file named methylation_kmplot will be generated in the working directory.
methy_kmplot(cancer,gene,palette='jco')cancer
cancer name likes "COAD".
gene
gene name likes "KLF7".
palette the color palette to be used for coloring or filling by groups. Allowed values include scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco".
Example
methy_kmplot("COAD","KLF7")Extract the TPM matrix of a specific type of cancer in TCGA.
get_tpm(cancer)cancer
cancer name likes "COAD".
Example
get_tpm("COAD")
#> Cancer Group TSPAN6 TNMD DPM1 SCYL3 C1orf112 FGR CFH FUCA2
#> TCGA-CM-4743-01A COAD Tumor 4.83 0.00 6.54 1.92 1.50 2.72 3.82 6.05
#> TCGA-D5-6931-01A COAD Tumor 6.58 1.73 6.70 3.26 3.42 3.11 3.97 6.31
#> TCGA-AA-A00A-01A COAD Tumor 5.93 1.03 6.20 2.90 2.12 2.99 3.24 6.82
#> TCGA-AD-A5EK-01A COAD Tumor 7.36 0.47 8.03 2.75 2.75 1.51 2.26 6.35
#> TCGA-A6-2680-01A COAD Tumor 6.90 1.73 6.66 2.55 3.01 2.79 2.88 6.02Extract the TPM matrix of a specific type of cancer with paired samples (n>20) in TCGA.
get_paired_tpm(cancer)cancer
cancer name likes "COAD".
Example
get_paired_tpm("COAD")
#> Cancer Group TSPAN6 TNMD DPM1 SCYL3 C1orf112 FGR CFH FUCA2
#> TCGA-CM-4743-01A COAD Tumor 4.83 0.00 6.54 1.92 1.50 2.72 3.82 6.05
#> TCGA-D5-6931-01A COAD Tumor 6.58 1.73 6.70 3.26 3.42 3.11 3.97 6.31
#> TCGA-AA-A00A-01A COAD Tumor 5.93 1.03 6.20 2.90 2.12 2.99 3.24 6.82
#> TCGA-AD-A5EK-01A COAD Tumor 7.36 0.47 8.03 2.75 2.75 1.51 2.26 6.35
#> TCGA-A6-2680-01A COAD Tumor 6.90 1.73 6.66 2.55 3.01 2.79 2.88 6.02Extract the clinical information of a specific type of cancer in TCGA.
get_meta(cancer)cancer
cancer name likes "COAD".
Example
get_meta("COAD")
#> Cancer event time age gender stage
#> TCGA-3L-AA1B COAD 0 5.13 61 F I
#> TCGA-4N-A93T COAD 0 0.27 67 M III
#> TCGA-4T-AA8H COAD 0 5.33 42 F II
#> TCGA-5M-AAT4 COAD 1 1.63 74 M IV
#> TCGA-5M-AAT6 COAD 1 9.67 41 F IV
#> TCGA-5M-AATE COAD 0 40.00 76 M II
#> TCGA-A6-2671 COAD 0 21.60 86 M IV
Extract the TMB matrix of all samples in TCGA.
get_tmb()Example
get_tmb()
#> TMB
#> TCGA-OR-A5J1-01A 0.70
#> TCGA-OR-A5J2-01A 0.83
#> TCGA-OR-A5J3-01A 0.27
#> TCGA-OR-A5J5-01A 8.53
#> TCGA-OR-A5J6-01A 0.77Extract the MSI matrix of all samples in TCGA.
get_msi()Example
get_msi()
#> MSI
#> TCGA-OR-A5J1 0.275
#> TCGA-OR-A5J2 0.324
#> TCGA-OR-A5J3 0.343
#> TCGA-OR-A5J5 0.522
#> TCGA-OR-A5J6 0.289Extract the promoter methylation information of all samples in TCGA.
get_methy()Example
get_methy()
#> $probe
#> probe gene
#> 1 cg26705472 A4GALT
#> 3 cg06339629 AADAT
#> 5 cg14239811 AADATExtract the immune cell ratio of all samples in TCGA.
get_immu_ratio()Example
get_immu_ratio()
#> B cells memory B cells naive Dendritic cells activated
#> TCGA-OR-A5LD-01A 0.0069 0.0000 0.0000
#> TCGA-OR-A5KO-01A 0.0685 0.0000 0.0844
#> TCGA-OR-A5LA-01A 0.0000 0.0117 0.0000
#> TCGA-OR-A5JW-01A 0.0133 0.0000 0.0258
#> TCGA-PA-A5YG-01A 0.0085 0.0056 0.0100
#> TCGA-OR-A5JD-01A 0.0146 0.0000 0.0093
Extract the immune score of all samples in TCGA.
get_immuscore()Example
get_immuscore()
#> B cells memory B cells naive Dendritic cells activated
#> TCGA-OR-A5LD-01A 0.0069 0.0000 0.0000
#> TCGA-OR-A5KO-01A 0.0685 0.0000 0.0844
#> TCGA-OR-A5LA-01A 0.0000 0.0117 0.0000
#> TCGA-OR-A5JW-01A 0.0133 0.0000 0.0258
#> TCGA-PA-A5YG-01A 0.0085 0.0056 0.0100
Return the sample summary of 33 types of cancer in TCGA.
get_cancers()Example
get_cancers()
#> Normal Tumor
#> ACC 0 79
#> BLCA 19 409
#> BRCA 113 1113
#> CESC 3 306
#> CHOL 9 35
#> COAD 41 473
#> DLBC 0 48
#> ESCA 13 185Return the sample summary of 15 types of cancer containing more than 20 paired samples in TCGA
get_paired_cancers()Example
get_paired_cancers()
#> Normal Tumor
#> BLCA 19 19
#> BRCA 113 113
#> COAD 41 41
#> ESCA 13 13
#> HNSC 43 43
#> KICH 25 25sessionInfo()
#> R version 4.3.1 (2023-06-16 ucrt)
#> Platform: x86_64-w64-mingw32/x64 (64-bit)
#> Running under: Windows 10 x64 (build 19044)
#>
#> Matrix products: default
#>
#>
#> locale:
#> [1] LC_COLLATE=Chinese (Simplified)_China.utf8 LC_CTYPE=Chinese (Simplified)_China.utf8
#> [3] LC_MONETARY=Chinese (Simplified)_China.utf8 LC_NUMERIC=C
#> [5] LC_TIME=Chinese (Simplified)_China.utf8
#>
#> time zone: Asia/Shanghai
#> tzcode source: internal
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] TCGAplot_0.99.0 testthat_3.2.0 ggpubr_0.6.0 ggplot2_3.4.3
#>
#> loaded via a namespace (and not attached):
#> [1] fs_1.6.3 matrixStats_1.0.0 bitops_1.0-7
#> [4] enrichplot_1.21.3 devtools_2.4.5 HDO.db_0.99.1
#> [7] httr_1.4.7 RColorBrewer_1.1-3 doParallel_1.0.17
#> [10] profvis_0.3.8 tools_4.3.1 backports_1.4.1
#> [13] utf8_1.2.3 R6_2.5.1 lazyeval_0.2.2
#> [16] GetoptLong_1.0.5 urlchecker_1.0.1 withr_2.5.1
#> [19] prettyunits_1.2.0 gridExtra_2.3 cli_3.6.1
#> [22] Biobase_2.61.0 scatterpie_0.2.1 survMisc_0.5.6
#> [25] yulab.utils_0.1.0 gson_0.1.0 DOSE_3.27.2
#> [28] sessioninfo_1.2.2 limma_3.57.9 rstudioapi_0.15.0
#> [31] RSQLite_2.3.1 generics_0.1.3 gridGraphics_0.5-1
#> [34] shape_1.4.6 car_3.1-2 dplyr_1.1.3
#> [37] GO.db_3.18.0 Matrix_1.6-1 waldo_0.5.1
#> [40] fansi_1.0.4 S4Vectors_0.39.2 abind_1.4-5
#> [43] lifecycle_1.0.3 whisker_0.4.1 yaml_2.3.7
#> [46] edgeR_3.99.0 carData_3.0-5 qvalue_2.33.0
#> [49] BiocFileCache_2.9.1 grid_4.3.1 blob_1.2.4
#> [52] promises_1.2.1 crayon_1.5.2 miniUI_0.1.1.1
#> [55] lattice_0.21-9 cowplot_1.1.1 KEGGREST_1.41.4
#> [58] pillar_1.9.0 knitr_1.44 ComplexHeatmap_2.17.0
#> [61] fgsea_1.27.1 rjson_0.2.21 codetools_0.2-19
#> [64] fastmatch_1.1-4 glue_1.6.2 ggfun_0.1.3
#> [67] data.table_1.14.8 remotes_2.4.2.1 fmsb_0.7.5
#> [70] vctrs_0.6.3 png_0.1-8 treeio_1.25.4
#> [73] gtable_0.3.4 rematch2_2.1.2 cachem_1.0.8
#> [76] xfun_0.40 mime_0.12 tidygraph_1.2.3
#> [79] survival_3.5-7 diffobj_0.3.5 pheatmap_1.0.12
#> [82] iterators_1.0.14 KMsurv_0.1-5 statmod_1.5.0
#> [85] interactiveDisplayBase_1.39.0 ellipsis_0.3.2 nlme_3.1-163
#> [88] ggtree_3.9.1 usethis_2.2.2 bit64_4.0.5
#> [91] filelock_1.0.2 GenomeInfoDb_1.37.6 rprojroot_2.0.3
#> [94] colorspace_2.1-0 BiocGenerics_0.47.0 DBI_1.1.3
#> [97] mnormt_2.1.1 tidyselect_1.2.0 processx_3.8.2
#> [100] bit_4.0.5 compiler_4.3.1 curl_5.1.0
#> [103] xml2_1.3.5 desc_1.4.2 shadowtext_0.1.2
#> [106] checkmate_2.2.0 scales_1.2.1 psych_2.3.9
#> [109] callr_3.7.3 rappdirs_0.3.3 stringr_1.5.0
#> [112] digest_0.6.33 rmarkdown_2.25 XVector_0.41.1
#> [115] htmltools_0.5.6 pkgconfig_2.0.3 dbplyr_2.3.4
#> [118] fastmap_1.1.1 rlang_1.1.1 GlobalOptions_0.1.2
#> [121] htmlwidgets_1.6.2 shiny_1.7.5 tinyarray_2.3.1
#> [124] farver_2.1.1 zoo_1.8-12 jsonlite_1.8.7
#> [127] BiocParallel_1.35.4 GOSemSim_2.27.3 RCurl_1.98-1.12
#> [130] magrittr_2.0.3 GenomeInfoDbData_1.2.10 ggplotify_0.1.2
#> [133] patchwork_1.1.3 munsell_0.5.0 Rcpp_1.0.11
#> [136] ape_5.7-1 viridis_0.6.4 stringi_1.7.12
#> [139] pROC_1.18.4 ggraph_2.1.0 brio_1.1.3
#> [142] zlibbioc_1.47.0 MASS_7.3-60 AnnotationHub_3.9.2
#> [145] plyr_1.8.8 org.Hs.eg.db_3.18.0 pkgbuild_1.4.2
#> [148] parallel_4.3.1 HPO.db_0.99.2 ggrepel_0.9.3
#> [151] survminer_0.4.9 Biostrings_2.69.2 graphlayouts_1.0.1
#> [154] splines_4.3.1 circlize_0.4.15 locfit_1.5-9.8
#> [157] ps_1.7.5 igraph_1.5.1 ggsignif_0.6.4
#> [160] reshape2_1.4.4 stats4_4.3.1 pkgload_1.3.3
#> [163] BiocVersion_3.18.0 evaluate_0.22 BiocManager_1.30.22
#> [166] foreach_1.5.2 tweenr_2.0.2 httpuv_1.6.11
#> [169] tidyr_1.3.0 purrr_1.0.2 polyclip_1.10-6
#> [172] km.ci_0.5-6 clue_0.3-65 ggforce_0.4.1
#> [175] broom_1.0.5 xtable_1.8-4 tidytree_0.4.5
#> [178] roxygen2_7.2.3 MPO.db_0.99.7 rstatix_0.7.2
#> [181] later_1.3.1 viridisLite_0.4.2 tibble_3.2.1
#> [184] clusterProfiler_4.9.4 aplot_0.2.2 forestplot_3.1.3
#> [187] memoise_2.0.1 AnnotationDbi_1.63.2 IRanges_2.35.2
#> [190] cluster_2.1.4