- Link to challenge: https://www.kaggle.com/c/bkai-igh-neopolyp/
- This dataset contains 1200 images (1000 WLI images and 200 FICE images) with fine-grained segmentation annotations. The training set consists of 1000 images, and the test set consists of 200 images. All polyps are classified into neoplastic or non-neoplastic classes denoted by red and green colors, respectively. This dataset is a part of a bigger dataset called NeoPolyp.
The code is structured as following:
project
│ README.md
└───data
│ └───test
| | | test1.jpeg
| | | test2.jpeg
| | | ...
│ │
│ └───train
│ │
│ └───train_gt
└───logs
│
└───models
│
└───loss
|
└───res
|
└───weights
| config.py
| dataset.py
| main.py
| mask2csv.py
| test.py
data : Where you put the training data and ground truth. There are three sub-folders test, train and train_gt in this folder. Please download data from https://www.kaggle.com/c/bkai-igh-neopolyp/data , and put the images in the corresponding folders ( only the images).
logs: You should save the training log of the models here.\
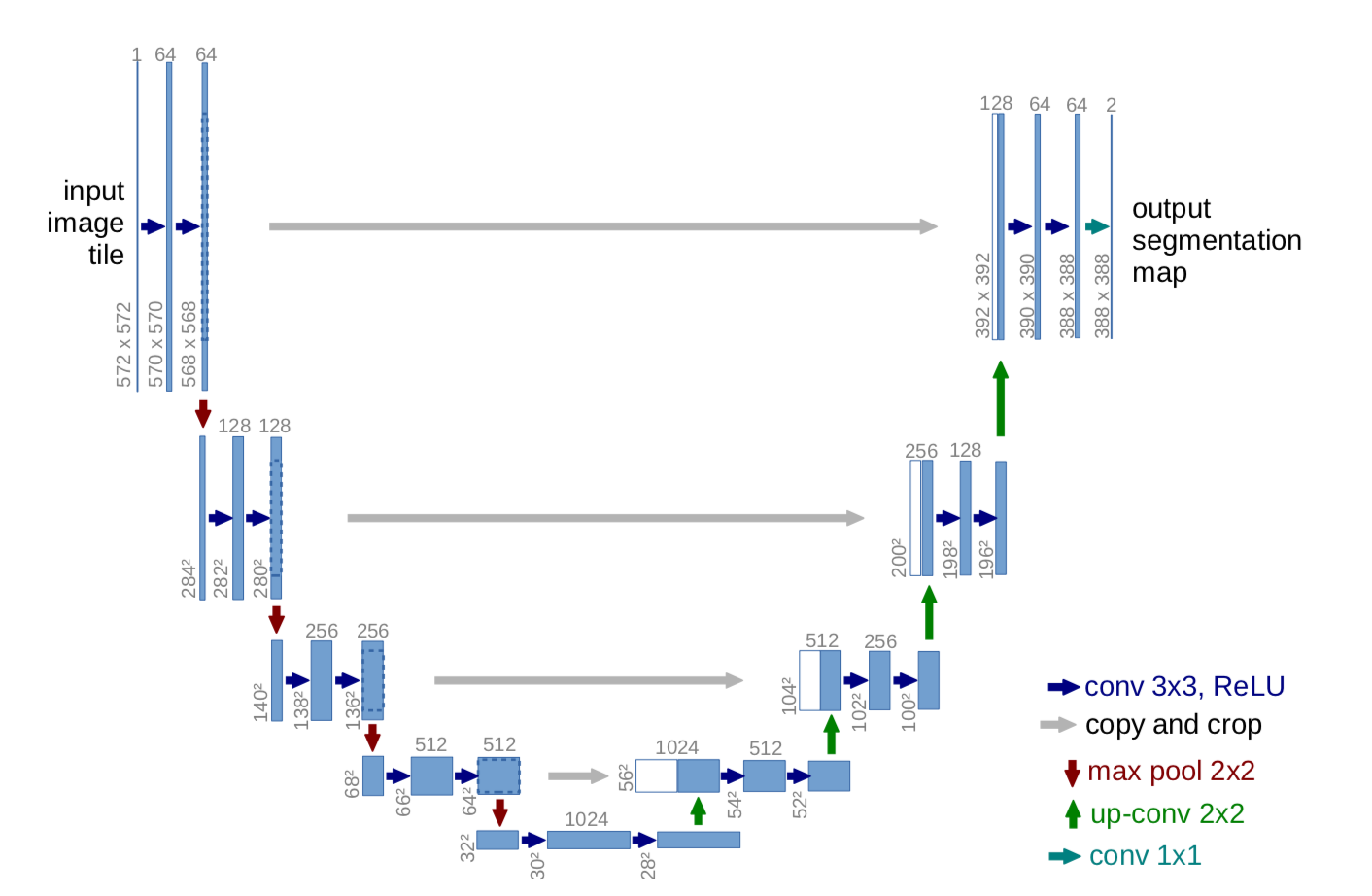
models : Code implementation of the models. Current supported models are: Unet and Attention Unet , and with three diferent backbones: Mobilnetv2,Efficientnet B0 and VGG-16.
res : inference results are saved here.
loss : implementation of loss functions
weights : weights are saved here.
config.py : Config for paths and model parameters
dataset.py : dataset-related utility functions
main.py : main training script.
mask2csv : convert from image to csv for kaggle submission
To train the model for polyps segmentation , please change the following parameters in config.py
IMAGE_PATH = '/media/HDD/bkai/data/train'
MASK_PATH = '/media/HDD/bkai/data/train_gt'
WIDTH = 256
HEIGHT = 256
BATCH_SIZE = 8
MODEL_SELECTION = "mb2_unet"
IMAGE_PATH : path to the train dataset directory
MASK_PATH : path to the train ground truth dataset directory
TEST_PATH : path to the test dataset directory
WIDTH and HEIGHT : input size of the model. Currently only supports 256x256. You can make it dynamic by making modifications to models in the folder models.
BATCH_SIZE: batch size
MODEL_SELECTION: Currently supports: unet (Unet with VGG-16 backbone), unet_eff0 (Unet with Efficientnet B0 backbone),mb2_unet (Unet with MobilenetV2 backbone) ,att_unet (Attention Unet), att_unet_mb2 and unet_att_eff0.
Then simply run main.py
To run inference , please change the following parameters in config.py
TEST_PATH = '/media/HDD/bkai/data/test'
WEIGHT_PATH = "/media/HDD/bkai/weights/mobinetv2_unet.h5"
SAVE_FOLDER = "/media/HDD/bkai/res"
SHOW_IMAGE = True
TEST_PATH : path to the test dataset directory
WEIGHT_PATH : path to the trained weight
SAVE_FOLDER : folder to save the inference result
SHOW_IMAGE: If you want to visualize the result one by one and show overlay result , please set to True. If you want to save the segmentation result , set to False. Please note that the visualization result if SHOW_IMAGE= True won't be smooth since it's the original one-hot map of size 256x256x3 and has not been resized to the original input size yet.
To convert result to .csv , please change the following parameters in config.py
MASK_RES_PATH = '/media/HDD/bkai/res'
MASK_RES_PATH : path to the inference result directory
- Information about Unet models are located at
models/Unet.py,models/Unetefficientb0.py,models/Unetmobilev2.py.
- Information about Attention Unet models are located at
models/AttentionUnet.py,models/AttentionUnetefficientb0.py,models/AttentionUnetmobilev2.py.
We need segent 3 classes:
-
0 if the pixel is part of the image background (denoted by black color);
-
1 if the pixel is part of a non-neoplastic polyp (denoted by green color);
-
2 if the pixel is part of a neoplastic polyp (denoted by red color).
See function convert2TfDataset in dataset.py.
The output of the model will be of size H,W,3, with 3 channels corresponding to 3 classes. The output will then be converted to corresponding RGB image
colors = np.array([[ 0, 0, 0],
[ 0, 255, 0],
[ 255, 0, 0]])
p = cv2.resize(p.astype(np.float32),(w,h), cv2.INTER_CUBIC) #H,W,3
p = np.argmax(p,axis=-1) # 995,1280
rgb = np.zeros((*p.shape, 3))
for label, color in enumerate(colors):
rgb[p == label] = color