Integrated DLO HiC analyze pipeline.
This package is only support UNIX-like system.
Recommend install and manage requirements with conda and mamba, just use following commands:
$ git clone https://github.com/Nanguage/DLO-HiC-Tools.git $ cd DLO-HiC-Tools/ $ conda install mamba -c conda-forge # skip this line if mamba is installed $ mamba env create -f environment.yml $ conda activate dlo-hic-tools $ python setup.py install
You can also using the docker, pull the dlohic image:
$ docker pull nanguage/dlohic
Run a container, with mount current directory in file system to the '/data' in the container:
$ docker run -v $PWD:/data -ti nanguage/dlohic:latest
DLO-HiC-Tools is an integrated automatic pipeline for DLO-HiC data analyze. Before we start, you should install package and some dependency. see installation.
DLO-HiC-Tools implemented some tools for DLO-HiC data analyze and quality control. You can list them and get more usage detail by dlohic command:
$ dlohic --help Usage: dlohic [OPTIONS] COMMAND [ARGS]... DLO HiC Tools command line interface. Options: --log-file TEXT The log file, default output to stdout. --debug / --no-debug If debug will print all information, default False. --version Show the version and exit. --help Show this message and exit. Commands: PET_span_dist Count the distribution of PET span. bedpe2pairs Transform BEDPE format file to pairs format,... build_bedpe Build bedpe file from fastq files. chr_interactions Count the interactions between chromosomes. extract_PET Extract the PETs sequences on both sides of... extract_fragments Extract all DNA fragments from fasta file,... fragment_length_dist Draw the distribution figure(kde/box plot),... gen_qc_report Generate pipeline quality control report. infer_adapter Inference adapter sequence, and plot the... interactions_qc Count ratio of: inter-chromosome... noise_reduce Remove noise in DLO-HiC data. pipeline Generate integrated main processes... plot_contact_map Draw the Hi-C contact map. remove_redundancy Remove the redundancy within pairs.
DLO-HiC-Tools implemented the pipeline with Snakemake. If you want generate the HiC matrix and the quality control report(see example) at once, you can use this pipeline.
The pipeline contain two files, a Snakemake file and a configuration file.
Firstly, generate them by dlohic pipeline command. It will generate the necessary files under your working directory.
$ dlohic pipeline dlo_hic.tools.helper.pipeline INFO @ 08/21/18 21:40:30: Generate config file at ./pipeline_config.ini dlo_hic.tools.helper.pipeline INFO @ 08/21/18 21:40:30: Generate pipeline (Snakemake file) at ./Snakefile $ ls pipeline_config.ini Snakefile
Then edit the pipeline_config.ini file, just fill the sections with it's comment information. And you can reference the example data and the configuration file, see here.
After all necessary information wrote in to the configuration file, You can run the pipeline.
$ snakemake -j 16
The -j parameter indicate the number of jobs, it depend on your cpu cores number. Or, if you use the cluster like pbs system, you can run like this:
$ snakemake --cluster qsub -j 16
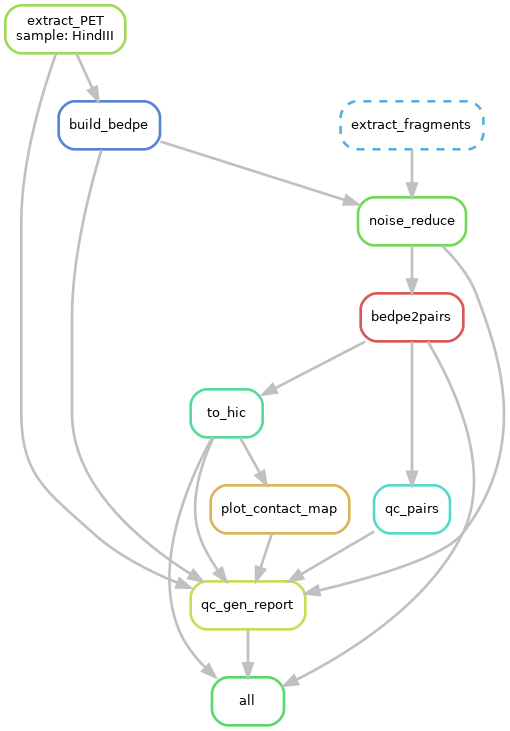
In addition, you can visualize the pipeline(need Graphviz installed):
$ snakemake all --dag | dot -Tpng > pipeline.png
More information about the Snakemake, please read it's document.
The pipeline can generate two kind of result matrix format, .hic and .cool. Although there are some contact maps in the HTML report, but if you want to browse the contact at finer resolution. You should use the software like: