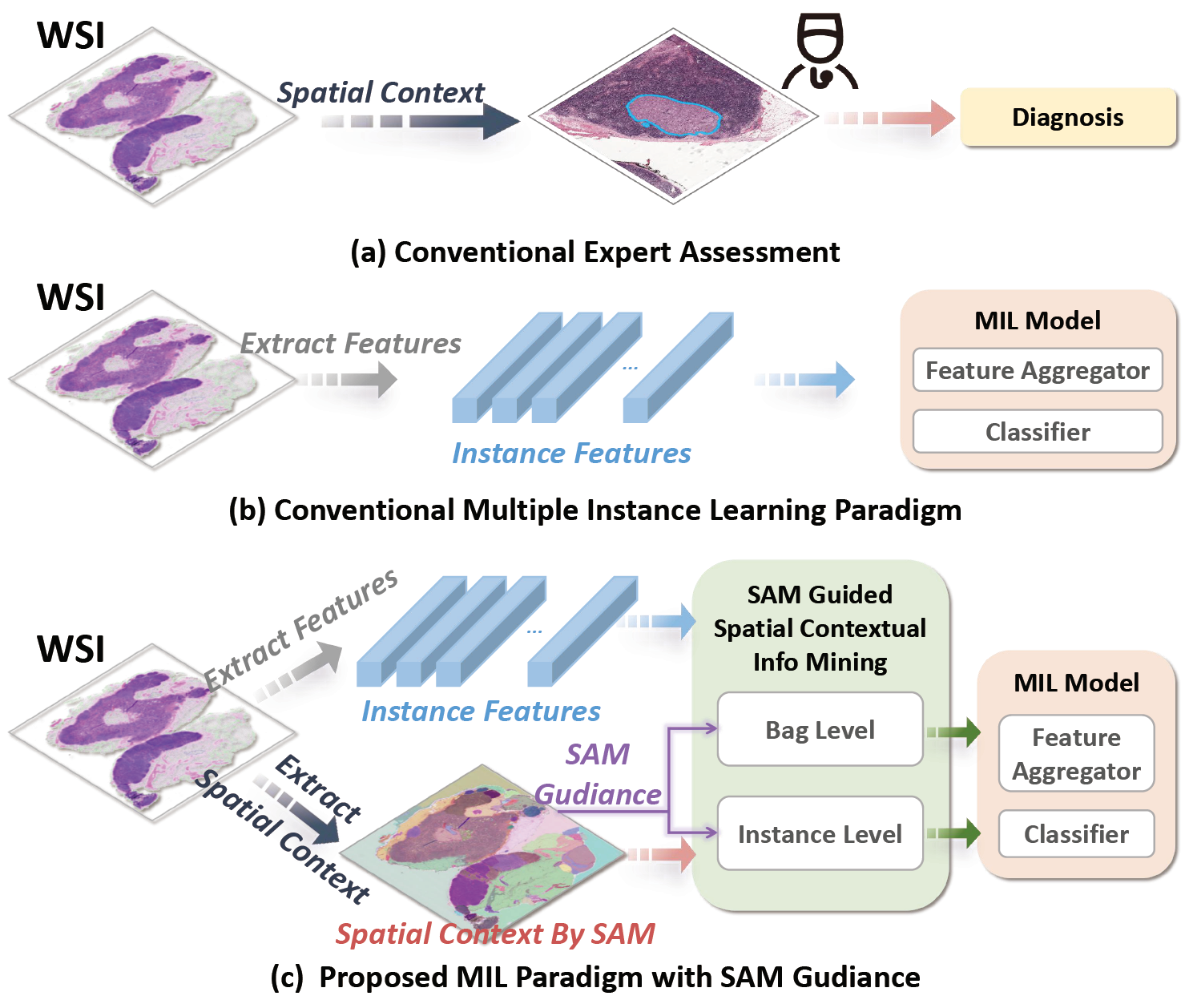
Official repository of "SAM-MIL: A Spatial Contextual Aware Multiple Instance Learning Approach for Whole Slide Image Classification", ACM Multimedia 2024. [arXiv] [PDF]
- Add the code for the training and testing of the model.
- Add the code for the preprocessing of the datasets.
- Add the code for the visualization of the results.
- Add the extracted features of the datasets(Camelyon16, TCGA-NSCLC).
- Improving README document.
- Improving the code structure.
We used the Segment Anything Model from the official repository to implement visual segmentation of WSIs.
- The weights used in the experiment can be found here.
- We used sam_vit_h as the backbone model for the segmentation task.
The preprocessing code can be found in the WSI_preprocess folder.
-
We adjusted the preprocessing steps in the CLAM repository. We made modifications to the original data preprocessing.
-
PLIP model and weight can be found in this.
Camelyon16:
python 01_create_patches_and_sam_segment.py --source '/path/to/your/WSI/folder' --save_dir 'path/to/save/patches' --patch_size 512 --step_size 512 --preset 'bwh_biopsy.csv' --seg --patch --stitch --use_sam --sam_checkpoint 'path/to/sam_weights.pth'TCGA-NSCLC:
python 01_create_patches_and_sam_segment.py --source '/path/to/your/WSI/folder' --save_dir '/path/to/save/patches' --patch_size 512 --step_size 512 --preset 'tcga.csv' --seg --patch --stitch --use_sam --sam_checkpoint '/path/to/sam_weights.pth'Camelyon16:
python 02_extract_features_and_group_feature.py --data_h5_dir '/path/to/patches' --data_slide_dir '/path/to/WSIs' --data_segment_dir '/path/to/segments' --csv_path '/path/to/process_list_autogen.csv' --feat_dir '/path/to/save/features' --use_sam --patch_size 512 --batch_size 512 --target_patch_size 224 --slide_ext .tifTCGA-NSCLC:
python 02_extract_features_and_group_feature.py --data_h5_dir '/path/to/patches' --data_slide_dir '/path/to/WSIs' --data_segment_dir '/path/to/segments' --csv_path '/path/to/process_list_autogen.csv' --feat_dir '/path/to/save/features' --use_sam --patch_size 512 --batch_size 512 --target_patch_size 224 --slide_ext .svsFrom features(.h5) to our model input:
python extract_features_from_h5.py --data_feat_h5_dir '/path/to/h5/features' --data_slide_dir '/path/to/WSIs' --data_segment_dir '/path/to/segments' --csv_path '/path/to/process_list_autogen.csv' --feat_dir '/path/to/save/features' --patch_size 512 --slide_ext .tif/.svsFrom features(.pt) to our model input:
python extract_features_from_pt.py --data_feat_pt_dir '/path/to/pt/features' --data_slide_dir '/path/to/WSIs' --data_segment_dir '/path/to/segments' --csv_path '/path/to/process_list_autogen.csv' --feat_dir '/path/to/save/features' --patch_size 512 --slide_ext .tif/.svsSome code snippets about PLIP feature are shown below:
extract_features_fp.py:
model = PLIP()
n_parameters = sum(p.numel() for p in model.parameters() if p.requires_grad)
mean, std = (0.48145466, 0.4578275, 0.40821073), (0.26862954, 0.26130258, 0.27577711)
eval_transforms = transforms.Compose([transforms.ToTensor(), transforms.Normalize(mean = mean, std = std)])
models/plip.py
from transformers import CLIPVisionModelWithProjection
class PLIP(torch.nn.Module):
def __init__(self):
super(PLIPM,self).__init__()
self.model = model = CLIPVisionModelWithProjection.from_pretrained("vinid/plip")
def forward(self, input):
return self.model(batch_input).image_embeds
The arguments for training can be found in options.py.
Train the model:
python main.py --project=your_project --datasets=camelyon16/tcga --dataset_root=/path/to/your/dataset --model_path=/path/to/save/model --cv_fold=5 --title=your_title --model=sam --sam_mask --mask_non_group_feat --mask_by_seg_area --backbone=attn --model_ema --mrh_sche --seed=2021 --mask_ratio=0.9 --select_mask --num_group=5 --group_alpha=0.5 --consistency_alpha=1000 --no_merge --num_workers=0 --persistence --wandbIf you find SAM-MIL useful in your research, please consider citing the following paper:
@article{fang2024sam,
title={SAM-MIL: A Spatial Contextual Aware Multiple Instance Learning Approach for Whole Slide Image Classification},
author={Fang, Heng and Huang, Sheng and Tang, Wenhao and Huangfu, Luwen and Liu, Bo},
journal={arXiv preprint arXiv:2407.17689},
year={2024}
}