Estimate bisulfite conversion frequency based on Sanger sequencing data of converted PCR products.
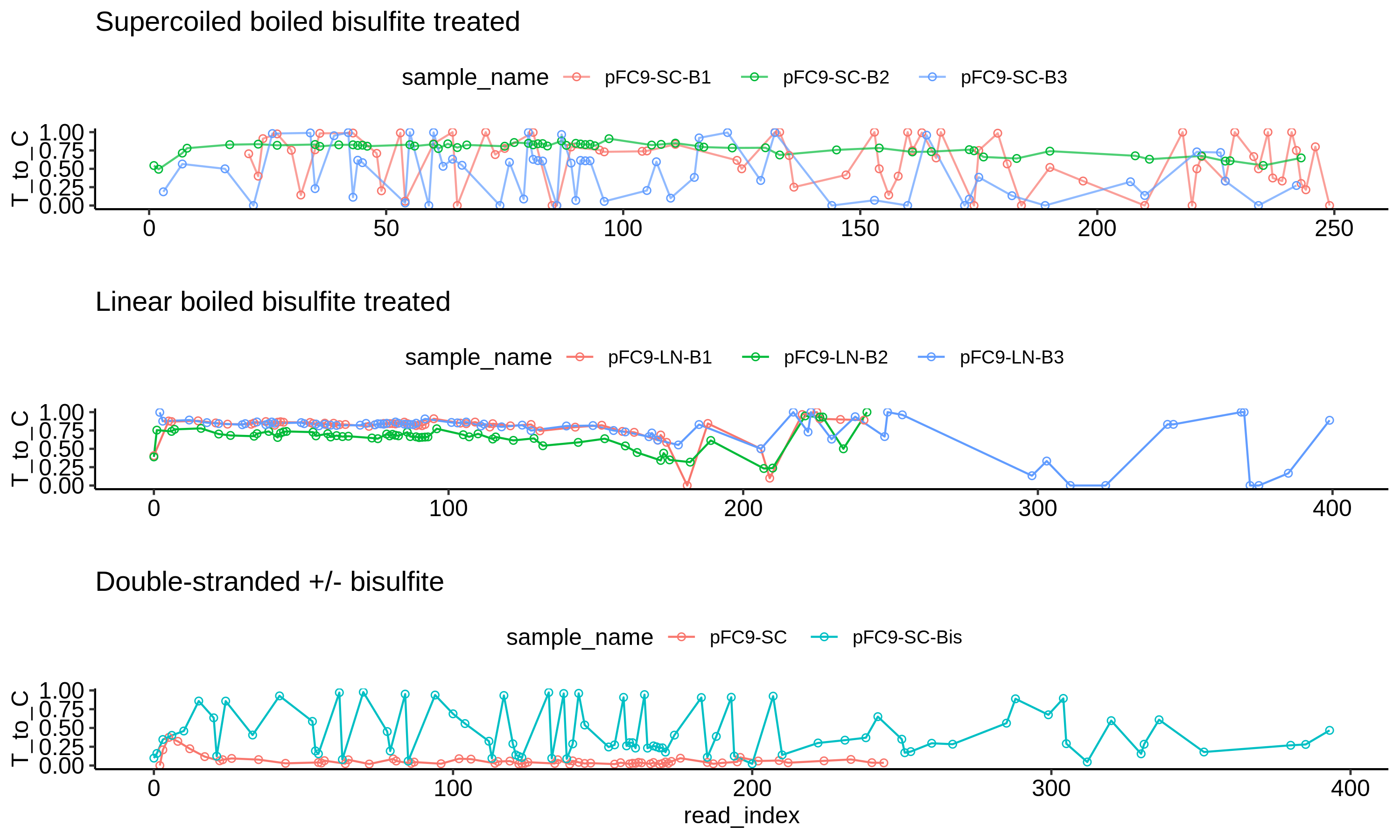
Non-denaturing bisulfite treatment converts single stranded cytosine nucleotides to thyamine nucleotides. Treating a homogenous plasmid substate with bisulfite and PCR amplifying the resulting products will give you a new population of sequences with some degree of C -> T conversion.
Subjecting this population to Sanger sequencing allows us to estimate the efficiency of conversion by comparing relative C and T RFU values at potential conversion sites. The higher the ratio of T to C signal the greater the conversion at that site.
This requires processing, aligning and visualizing raw Sanger trace data. This Snakemake based workflow is designed to do just that.
- Install snakemake
- Run
snakemake -j 1 --configfile workflow/config.yml --use-condafrom the root directory of the repo.