Code for a project investigating the effective dimensionality of deep neural networks, and its relationship to their ability to predict neural activity in visual cortex. Described in the paper: High-performing neural network models of visual cortex benefit from high latent dimensionality
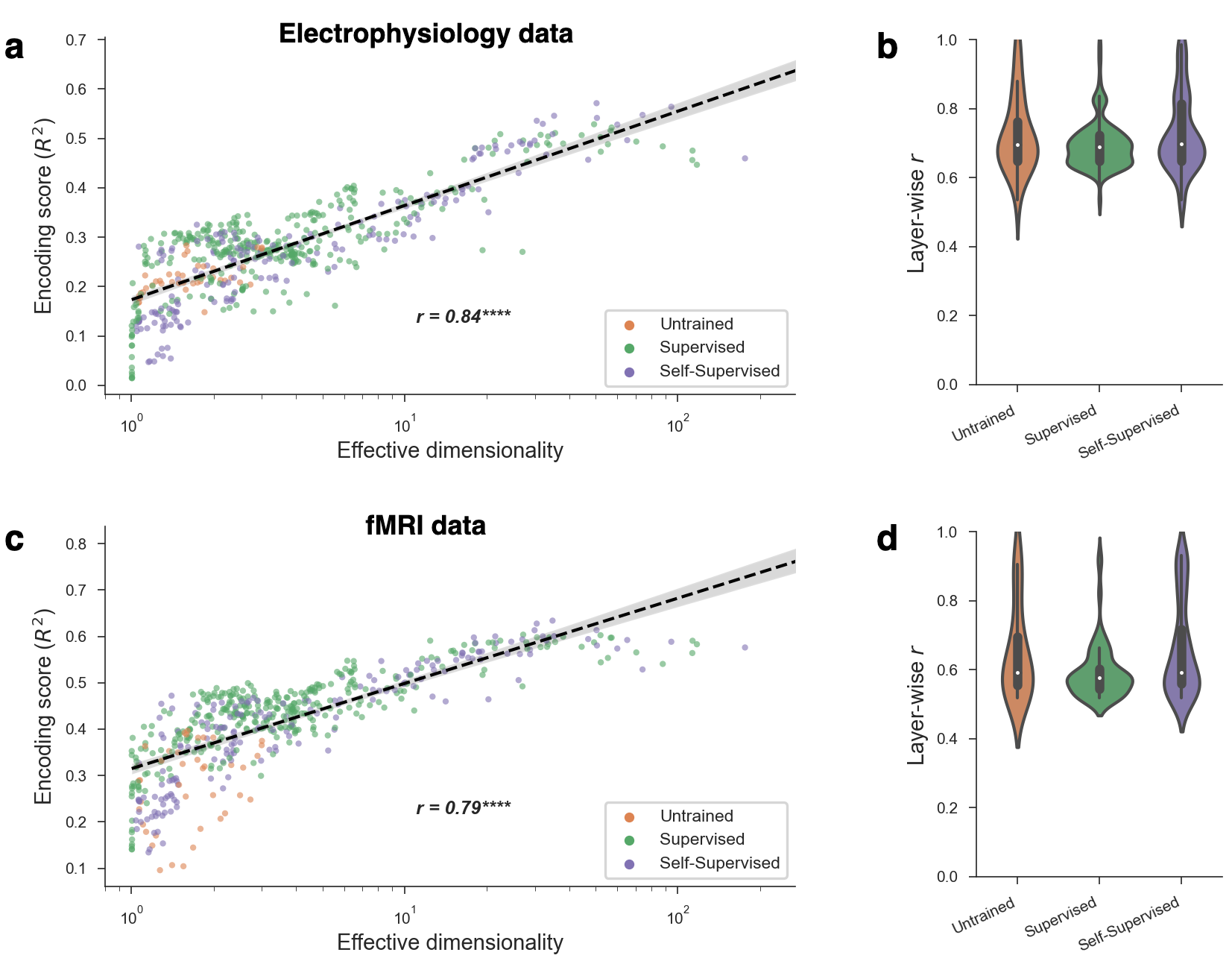
Geometric descriptions of deep neural networks (DNNs) have the potential to uncover core principles of computational models in neuroscience, while abstracting over the details of model architectures and training paradigms. Here we examined the geometry of DNN models of visual cortex by quantifying the latent dimensionality of their natural image representations. The prevailing view holds that optimal DNNs compress their representations onto low-dimensional manifolds to achieve invariance and robustness, which suggests that better models of visual cortex should have low-dimensional geometries. Surprisingly, we found a strong trend in the opposite direction—neural networks with high-dimensional image manifolds tend to have better generalization performance when predicting cortical responses to held-out stimuli in both monkey electrophysiology and human fMRI data. These findings held across a diversity of design parameters for DNNs, and they suggest a general principle whereby high-dimensional geometry confers a striking benefit to DNN models of visual cortex.
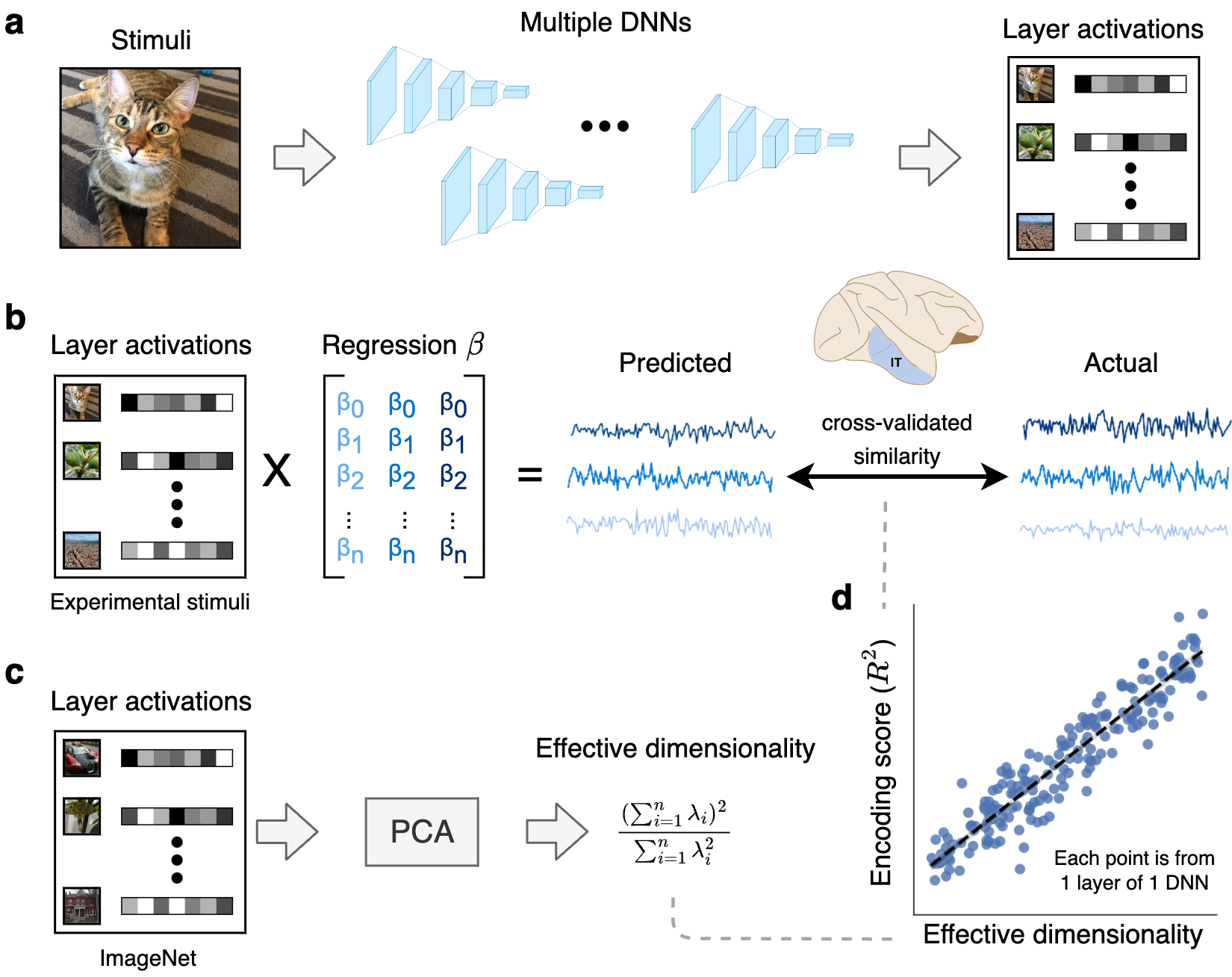
Method for comparing latent dimensionality with encoding performance for neural data. a. Layer activations were extracted from a large bank of DNNs trained with different tasks, datasets, and architectures. b. Using these layer activations as input, we fit linear encoding models to predict neural activity elicited by the same stimuli in both monkey and human visual cortex. We used cross-validation to evaluate encoding performance on unseen stimuli. \textbf{c.} To estimate the effective dimensionality of our models, we ran principal component analysis on layer activations obtained from a large dataset of naturalistic stimuli (specifically, 10,000 images from the ImageNet validation set). \textbf{d.} These analyses allowed us to examine the empirical relationship between effective dimensionality and linear encoding performance across a diverse set of DNNs and layers. DNN = deep neural network, PCA = principal component analysis.
Relationship between effective dimensionality and encoding performance. a. The encoding performance achieved by a model scaled with its effective dimensionality (Pearson
The code has been tested on Python 3.7.x with the following packages:
brain-scorepackage linkmodel-toolspackage linkcandidate-modelspackage linkjupyter-lab(for the figure generation)
The above is not an exaustive list. There are number of additional required packages (e.g. pytorch, tensorflow, xarray), which should be installed automatically as dependencies of those in the above list if you're using pip. After that, just try running the scripts following the commands below and install any remaining packages that Python complains are missing.
To reproduce the results in the main manuscript, the following scripts must be run from the root project directory with the indicated command line arguments.
To compute the effective dimensionalities of all models:
python -m scripts.compute_eigenspectra --dataset imagenet
To compute the encoding performance of all models:
python -m scripts.fit_encoding_models --no_pooling
To compute transfer classification performance on ImageNet-21k (please reach out to me to receive the dataset):
python -m scripts.compute_nshot --dataset imagenet21k --data_dir [directory to imagenet21k dataset] --classifier prototype
To compute model projection distances on ImageNet-21k (please reach out to me to receive the dataset):
python -m scripts.compute_projection_distances --dataset imagenet21k --data_dir [directory to imagenet21k dataset]
Each of these scripts will generate one or more *.csv file in the results/ directory.
To reproduce the figures in the main manuscript, run the jupyter notebooks in the figures/manuscript/ directory. These read from the generated *.csv result files.
Some code blocks generate figures from the Appendix and will rely on results files not created in the previous section (e.g. ZCA-transformed feature results). To generate those Appendix figures as well, run the other relevant scripts in the scripts/ directory (scroll down to the bottom of the files to see the required arguments). Don't hesitate to reach out to me if you run into issues during this process.