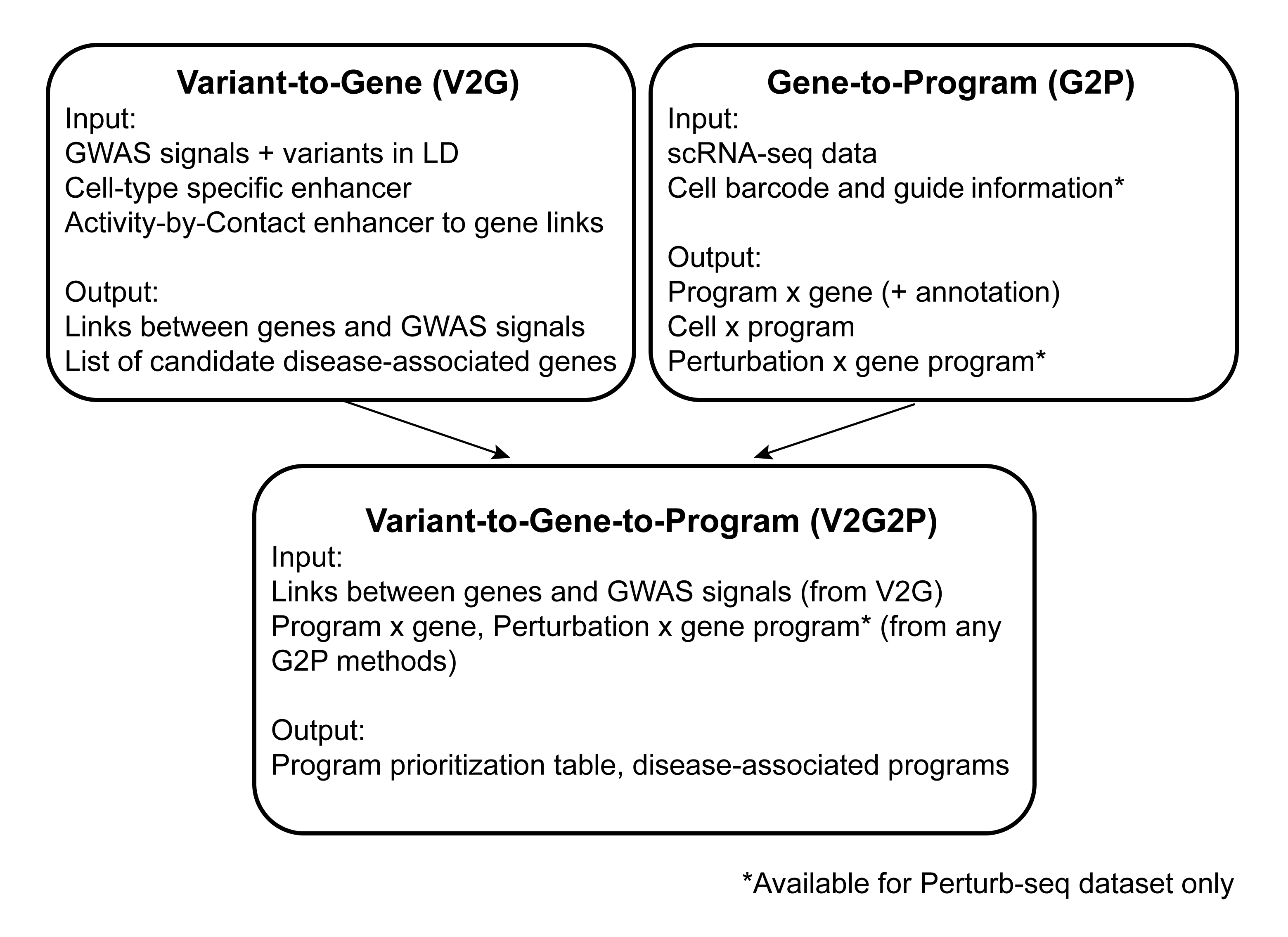
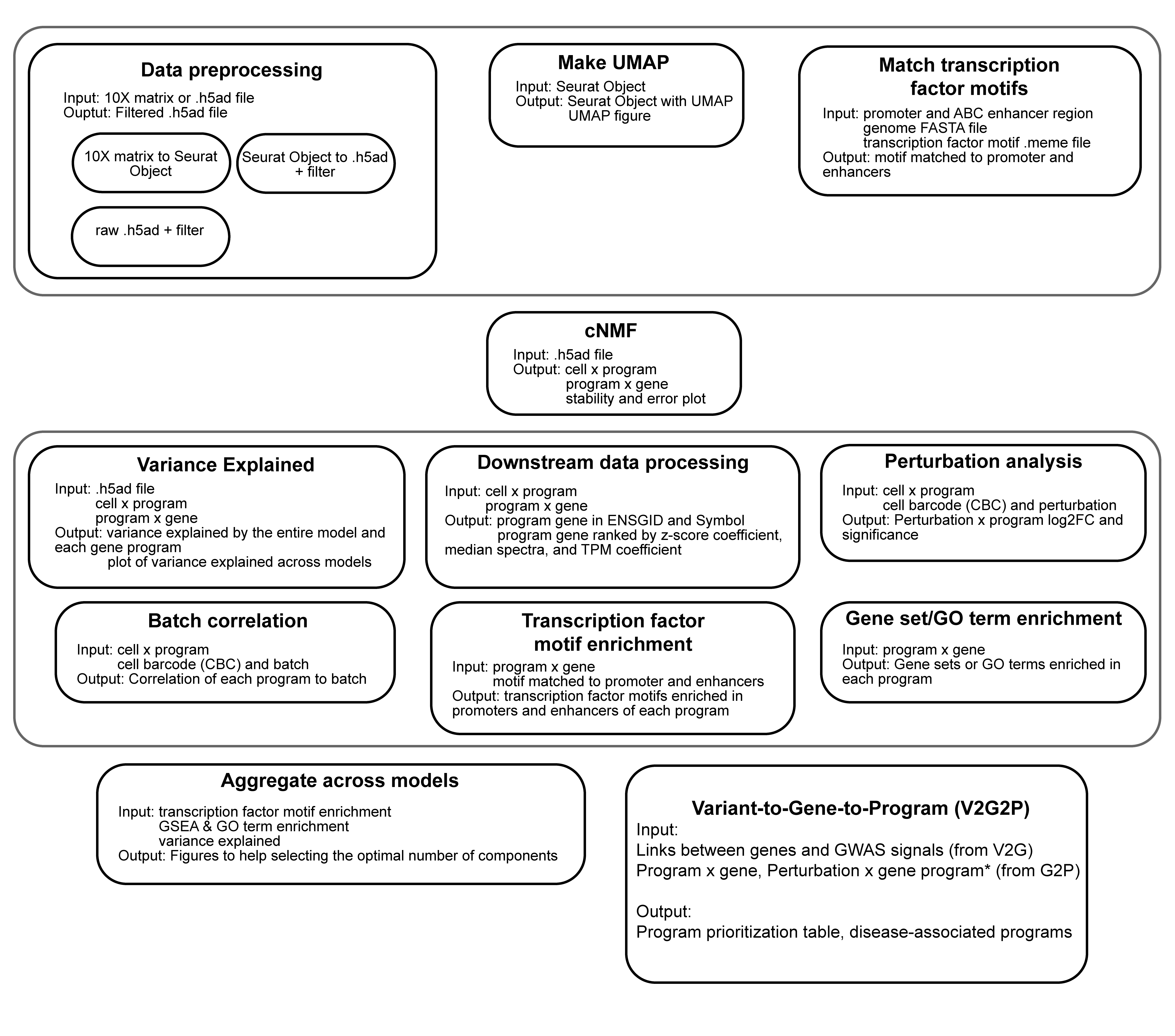
A pipeline to connect GWAS variants to genes to disease-associate gene programs. This pipeline uses snakemake and cNMF from Kotliar et al. The V2G2P approach could be applied to any GWAS studies with the correct cell type(s).
The V2G2P approach has three components: V2G, G2P, and V2G2P enrichment test. Each one works as a stand-alone pipeline. Together these three components are essential for V2G2P. Below is an overview of the relations among the three steps:
The V2G pipeline is linked here. Author: Rosa Ma.
Below is a figure showing the different modules and features within the G2P pipeline:
Install Snakemake and conda environment using conda:
conda env create -f conda_env/cnmf_env.yml
conda env create -f conda_env/cnmf_analysis_R.ymlcnmf_env contains snakemake. If you do not have snakemake installed already, you can activate the environment via conda activate cnmf_env, then run the pipeline.
Config file slots:
| field | meaning |
|---|---|
| total_workers | Number of processes to run in parallel |
| seed | A number to set seed for reproducibility |
| num_runs | Number of NMF run (recommend 100 for the actual data analysis, and 10 for testing the pipeline) |
| run_per_worker | Number of run assign to each process/worker to run in series (make sure that num_runs = total_workers * run_per_worker) |
| k | Number of components to factorize |
| thresholds | Threshold for filtering outlier components, 2 for no filtering, recommend 0.2 |
| analysisDir | Directory for numeric result outputs |
| figDir | Directory for figure outputs |
| dataDir | If using 10X scRNA-seq matrix as input, the directory with 10X files (file names must be features.tsv.gz, barcodes.tsv.gz, matrix.mtx.gz). This can be left blank if supplying the input matrix as an h5ad file. |
| input_h5ad_mtxDir | Path to input matrix in h5ad format. Please put the h5ad file in config["analysisDir"]/data/ folder. The snakemake pipeline will look for the input h5ad file from there. The file name must be the same as config["sampleName"] (e.g. config["analysisDir"]/data/{sample}.raw.h5ad, if you want to filter the data (remove noncoding genes, very lowly expressed genes, and cells with little genes detected) using this pipeline, please name the input file to '{sample}.raw.h5ad'. If not, use '{sample}.h5ad' to skip filtering). Leave this blank if supplying the input matrix in other formats. |
| scratchDir | Directory for cNMF pipeline to store temporary files |
| barcodeDir | (Necessary for Perturb-seq, optional for scRNA-seq without perturbation) Path to cell barcode / identifier tsv file, the sequence of appearance should be the same as in the input cell x gene matrix, and it must contain columns named "Gene" and "Guide" if doing Perturb-seq |
| motif_meme | MEME file for matching sequence to motifs |
| fasta_file | Fasta file to find sequences based on coordinates |
| promoter_fimo_formatted | (Optional) Path to promoter motif matches from FIMO output |
| enhancer_fimo_formatted | (Optional) Path to ABC enhancer motif matches from FIMO output |
| ABC_enhancers | Coordinates for ABC enhancers for the approapriate cell type |
| PoPS_raw_featureDir (not used in this version) | Directory for storing feature txt files (one row per gene in ENSGID) for PoPS input |
| PoPS_gene_annotation_path (not used in this version) | Gene annotation file for PoPS pipeline |
| magma_dir (not used in this version) | Path to MAGMA results for PoPS pipeline input |
| magma_prefix (not used in this version) | Desired magma_prefix for pointing to files in magma_dir |
| PoPS_control_features (not used in this version) | A list of features that serves as controls for the PoPS pipeline |
| PoPS_features_metadata_path (not used in this version) | PoPS external feature's annotation |
| pipelineDir (not used in this version) | The directory to this snakemake pipeline |
| K_spectra_threshold_table (Optional) | Table used for specifying spectra cut-off for each K tested in the pipeline. default: 0.2 for all K |
| GWAS_traits | GWAS traits to conduct V2G2P |
| sampleName | Name for this run |
| organism | Organism of the sample (human or mouse only) |
| cNMF_gene_selection | "all_genes" for using all expressed genes as input, "top3000VariableGenes" for using the top 3,000 most variable genes as an input, the number of top variable genes can be adjusted by the user, e.g. "top2000VariableGenes" for using the top 2,000 most variable genes. If using "all_genes", the user must specify the gene names in a .txt file in one column, stored at config["analysisDir"]/data/ with file name: {sample}.h5ad.all.genes.txt |
| Perturb-seq | True for an Perturb-seq experiment. The pipeline will generate additional results based on comparison between perturbation and control. False for any scRNA-seq experiment. |
| num_Genes_per_MAST_runGroup | (Optional, required for Perturb-seq datasets) Number of perturbations to be tested against control in one job submission. This is to speed up the code. Recommend 20. |
| numCtrls_for_MAST | (Optional, required for Perturb-seq datasets) Number of (randomly selected) controls to use in statstical test. Recommend 5000 or less for speed. |
| num_cells | Expected number of cells in the input dataset. It doesn't have to be exact. This is for optimizing memory requests and job scheduling. |
Note: please keep all optional items in the config file, even if it points to a non-existing path. The snakemake pipeline might return an error if they are not there.
conda activate cnmf_env
snakemake -n --configfile /path/to/config.json --quiet ## always recommend doing a dry runExecute the workflow locally via
snakemake --configfile /path/to/config.jsonPlease see the log.sh file in this github page for more examples.
For more snakemake usage and configuration, please visit snakemake documentation page.
The output files are in the folders specified in analysisDir and figDir fields in the config file.
Output can be found in config['analysisDir']/{cNMF_gene_selection}/{sampleName}/acrossK/
Output can be found in config['analysisDir']/{cNMF_gene_selection}/{sampleName}/K*/threshold_*/
topic.zscore_k_*.dt_*.txt
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
ENSG00000237491 3.6257507558702e-05 -0.000235942153872956 -0.000193839434908331 -0.000540485719839952 0.000552241385286034 -0.000201390791494423 8.75377887444021e-05 -8.26346157936536e-05 0.000256759855447459 -0.000339609165657141 2.97896083950046e-05 0.000687455576483113 -0.000151702242624839 0.000327691107098052 -0.000204158001546107 -0.000289447771145073 -0.000196239011791769 0.00154040438524839 6.64307968052208e-05 -0.000210747623307798 3.91082396297854e-05 3.28090173953066e-05 -6.82762061474105e-05 0.000346680760437194 -0.000434505149419527 3.07134546629071e-05 -4.66919527344807e-05 -9.84821214904951e-05 -0.000310237477830714 -0.000638909219613249 -0.000449261618986614 0.00236169789924395 -2.99388207930532e-05 0.000112124250801755 -0.000205873107158084 0.00118396446801699 -8.04675518574793e-05 -1.08022761998793e-06 -0.00032657718959049 0.000879090403025747 0.000711604342284466 4.58589731592956e-05 7.81209925208869e-05 -9.68548257900938e-05 -0.000261418751767653 -6.97565760230801e-05 -0.000364045128671641 -9.14872433022581e-05 0.00301861664150378 0.000487734728605933 0.000144017860764802 0.000204245920744823 -0.00028592571008198 0.000114994441178753 2.33365636877884e-05 0.00179981045907693 -0.000127734743936086 -0.000473005106126543 -0.000286177991598109 -0.000218404922934822 -0.000225282612194657 0.000522523214578263 0.000170376906142333 -0.00035889738665674 -0.0003926501366325 0.000275056020755212 -0.000486186756372187 0.000238462372482112 0.000175202698436504 -0.000229673808428174 -0.000700905188911717 0.000144637250219059 9.12354369735676e-05 -8.40999629381241e-05 -0.000118252777148798 0.000172813493783571 8.74358760916756e-05 0.000404696722921066 0.000132028345127144 9.64011060392208e-05 0.000227205136515923 -0.000243261558681986 -0.000214146639495842 -0.000362649397197894 -5.11403787774615e-05 0.000740349657857787 7.65313173554984e-05 -0.000274844444217957 -0.000381361969318532 -0.000177794399546524
ENSG00000228794 0.0003830319516136 0.000355691338179841 -1.05950306027163e-05 0.000127820490880465 -7.31291821168389e-05 -1.10277689952049e-05 0.000109005307931079 0.000518538576359656 0.000485105320360724 2.27368424675288e-05 -0.000230314274890742 -0.000150283422291734 -0.00010529458664589 7.70193256240283e-05 -2.8272520097087e-06 -0.000565786025411416 -8.75795335952311e-05 0.000222131397817275 4.08464600104762e-06 -8.84891928573324e-05 0.000129675002348178 0.000330994369629849 -3.49911315134635e-05 7.698047595147e-05 -0.000207698872887955 -0.000365335395047771 -0.000358755651660673 -0.000127650412950376 -0.000180414420617124 -0.00119329814081874 3.43609322673543e-05 -0.000797526438995552 -0.000148774288156748 0.000354855300060872 -0.000227170502101143 -0.000225235408025414 -0.000199515103928078 0.000238588352532944 3.37933029526634e-05 0.000126233561182807 -0.000652257621179978 0.000137670808631218 -5.25055940520106e-05 7.79270777152396e-06 -0.000290050479113386 -0.000253159097195554 -0.000156294235706472 0.000110566235414443 6.04551701827383e-05 0.00204524774593777 1.76794497794074e-05 0.000183176421857839 -0.000156802463557547 -0.000267998745215926 -7.89076033040562e-05 0.000544368144273017 -0.000111469393298824 -0.000328172768008264 7.83562504370004e-05 2.2862276787585e-05 5.58245262902203e-05 -0.000141937166498772 -9.16882192704192e-05 -2.50244445672456e-05 -0.000333419229756242 0.000571927351385279 -0.00109361661296798 -3.85078624884629e-06 2.45876064353904e-05 -4.28323617580336e-06 -6.44043276606002e-05 6.96526886733287e-05 -5.13316341023035e-05 0.000101058711721449 0.000447087628331816 -9.54830431057188e-05 -9.84285278180958e-05 0.000382407144128984 -6.47913289005252e-05 0.000154304069623889 0.000278680354427714 9.02708817726633e-05 -4.77518643121104e-05 2.22121399107156e-05 -0.000100486230733875 -0.000155911349936267 9.58525674405382e-05 9.44644886135765e-05 -0.000188089365111741 6.06376279784062e-05
ENSG00000188976 0.000350524557558168 0.00140440015571758 0.000283150852501125 -0.000197248748171782 -0.00037510091461753 -0.000105836646173528 -0.000286203456393242 -0.000239557697032912 -0.000248794066914784 -0.0012257052578368 -0.000988751089409712 -0.000305878236350561 -0.000717408393449572 -0.00135462020016997 0.000218844657463507 -5.69693856103151e-06 0.000292575981933451 0.000802881308037214 -0.000463570681085942 -0.000546871152910976 0.000428807831608 -0.000332801001901376 -0.000363867768313229 1.26751236270919e-05 -0.000613844522555879 0.000134192538576475 -0.000151844697479961 -0.000517247366515382 -0.000979696735403971 -0.00231643290337771 -0.000411060807246556 -0.000829614535615998 -0.000463595357467624 0.000211114060032754 -0.000459602973957588 -0.000385539380306828 3.26837996307634e-05 4.90582823595978e-05 -0.000275485781302146 -0.000913505079533773 -0.000389164082050203 9.33356980316987e-05 0.000114413872875212 0.000486612202137258 0.000192577452022132 8.01864355453836e-06 -0.000516070978454305 0.000643960764244882 -0.000219556345385713 0.00536865311335595 -0.000535178856867003 -0.000584067084769768 -0.000414121189304587 -0.000326010941366969 -0.00045331624134027 0.00104428295695003 -7.06825973624717e-05 -0.000582497714918622 0.00083933424178752 -0.000160149329772988 9.39739949391142e-05 -0.000650835878212008 -0.000184777484265797 -0.000294350147880196 -0.000596363795966926 0.00131283875271719 -0.00155652921516448 -0.000717653881449108 -0.000204400568012354 -0.000421267000410629 -0.000156226991563029 0.000141473267184267 -0.000433393032937874 -0.00018681415810335 0.00108084886240902 -0.000508499883366756 -0.000399452289109583 0.00101750008745467 0.000629843969832574 -5.40811730509978e-05 -0.000608716271934908 -0.000141709694152402 0.00128779179684845 0.000729006620348706 3.08031909026722e-05 0.000257931810362615 -0.000243090135199539 0.000937109035270549 -0.000497197904106814 6.54297394566199e-06
Column description:
Each column is a program. Each row is a gene.
*_ProgramSummary_k_*.dt_*.xlsx
ProgramID MaxBatchCorrelation nSigPerturbationsProgramUp nSigPerturbationsProgramDown nSigMotifsPromoter nSigMotifsEnhancer ProgramGenesZScoreCoefficientTop10 ProgramGenesTPMCoefficientTop10 ProgramGenesMedianSpectraTop10 ProgramGenesMedianSpectraZScoreTop10 ProgramGenesMotifsPromoter ProgramGenesMotifsEnhancer ProgramGenesZScoreCoefficientGOTermsTop10 ProgramGenesMedianSpectraZScoreGOTermsTop10
K90_1 0.009690118 43 105 5 HMMR,DLGAP5,HMGB2,CDC20,MIR17HG,ENSG00000260708,CKS2,CCNB2,CCNB1 COX3,COX2,ATP6,CYTB,ND3,EEF1A1,COX1,RPLP1,ND4 ENSG00000260708,ZNF280B,MIR17HG,SOX4,TOB2,ENSG00000261526,DLGAP5,HMGB2,TMF1 TOB2,ZNF280B,MIR17HG,C21orf91,PIK3R1,SOX4,REST,RAB22A,NXF1 CEBPZ,NFYB,FOXI1,NFYC,NFYA BP:GO:0051301:cell division,CC:GO:0000779:condensed chromosome, centromeric region,CC:GO:0000793:condensed chromosome,BP:GO:0000070:mitotic sister chromatid segregation,BP:GO:0000819:sister chromatid segregation,BP:GO:0098813:nuclear chromosome segregation,BP:GO:0140014:mitotic nuclear division,CC:GO:0005819:spindle,CC:GO:0000776:kinetochore,CC:GO:0000775:chromosome, centromeric region MF:GO:0140110:transcription regulator activity,BP:GO:0006366:transcription by RNA polymerase II,CC:GO:0000779:condensed chromosome, centromeric region,CC:GO:0000777:condensed chromosome kinetochore,CC:GO:0044451:nucleoplasm part,BP:GO:0051301:cell division,BP:GO:0006357:regulation of transcription by RNA polymerase II,CC:GO:0000775:chromosome, centromeric region,CC:GO:1990234:transferase complex,CC:GO:0000776:kinetochore
K90_2 0.065779043 27 527 10 PITX1,SRSF5,SRSF6,NEFH,SRSF7,HIC2,TOMM40,SLC30A1,ZFP36L2 COX2,COX3,ATP6,ND4,COX1,CYTB,ND1,RPS2,ND3 PITX1,SRSF5,SRSF6,SRSF7,NEFH,TOMM40,HIC2,KCNH2,ZFP36L2 NEFH,NRARP,SRSF7,HIC2,ELL,PITX1,TOMM40,PPP1R37,SRSF5 AP2D,ZIC4,ZF64A,NR1H4,THAP1,SP1,ZN320,RFX1,EGR4,AP2B CC:GO:0030687:preribosome, large subunit precursor,CC:GO:0030684:preribosome,BP:GO:0043484:regulation of RNA splicing,CC:GO:0016607:nuclear speck,CC:GO:0044452:nucleolar part,BP:GO:0000381:regulation of alternative mRNA splicing, via spliceosome,BP:GO:0042254:ribosome biogenesis,BP:GO:0050684:regulation of mRNA processing,BP:GO:0048024:regulation of mRNA splicing, via spliceosome,BP:GO:1903311:regulation of mRNA metabolic process MF:GO:0003723:RNA binding,BP:GO:0006405:RNA export from nucleus,BP:GO:0051254:positive regulation of RNA metabolic process,BP:GO:0071166:ribonucleoprotein complex localization,BP:GO:0071426:ribonucleoprotein complex export from nucleus,BP:GO:0045935:positive regulation of nucleobase-containing compound metabolic process,BP:GO:0051168:nuclear export,BP:GO:1902680:positive regulation of RNA biosynthetic process,BP:GO:1903508:positive regulation of nucleic acid-templated transcription,BP:GO:0006403:RNA localization
K90_3 0.059547855 15 612 TFRC,PICALM,NBAS,PIK3CB,OPA1,PCNT,LMNB1,PRKDC,MACF1 COX3,ATP6,COX2,ND4,EEF1A1,COX1,NPM1,CYTB,NCL TFRC,PICALM,OPA1,NBAS,MACF1,PIK3CB,PCNT,LMNB1,TOP2B OSBPL3,ZNF318,PCNT,ZNF217,SYNRG,LMNB1,DENND4C,PPP6R3,ZFX BP:GO:0051301:cell division,MF:GO:0045296:cadherin binding,MF:GO:0050839:cell adhesion molecule binding,BP:GO:0051052:regulation of DNA metabolic process,MF:GO:0008092:cytoskeletal protein binding,CC:GO:0015629:actin cytoskeleton,MF:GO:0003729:mRNA binding,CC:GO:0099080:supramolecular complex,CC:GO:0099081:supramolecular polymer,CC:GO:0005925:focal adhesion MF:GO:0003723:RNA binding,MF:GO:0000166:nucleotide binding,MF:GO:1901265:nucleoside phosphate binding,MF:GO:0005524:ATP binding,MF:GO:0032559:adenyl ribonucleotide binding,MF:GO:0030554:adenyl nucleotide binding,MF:GO:0035639:purine ribonucleoside triphosphate binding,MF:GO:0032553:ribonucleotide binding,MF:GO:0032555:purine ribonucleotide binding,MF:GO:0017076:purine nucleotide binding
Column description:
ProgramID: Program identifier
MaxBatchCorrelation: Maximum Pearson correlation with batch
nSigPerturbationsProgramUp: Number of significant perturbations that upregulates the program
nSigPerturbationsProgramDown: Number of significant perturbations that downregulates the program
nSigMotifsPromoter: Number of transcription factor motifs enriched at top 300 gene's promoter (top 300 genes ranked by z-score coefficient)
nSigMotifsEnhancer: Number of transcription factor motifs enriched at top 300 gene's ABC linked enhancers (top 300 genes ranked by z-score coefficient)
ProgramGenesZScoreCoefficientTop10: Top 10 gene names, ranked by z-score coefficient from cNMF
ProgramGenesTPMCoefficientTop10: Top 10 gene names, ranked by TPM from cNMF
ProgramGenesMedianSpectraTop10: Top 10 gene names, ranked by raw weights from cNMF
ProgramGenesMedianSpectraZScoreTop10: Top 10 gene names, ranked by raw weight's z-score for each gene across programs
ProgramGenesMotifsPromoter: Transcription factor motifs enriched at top 300 gene's promoter (top 300 genes ranked by z-score coefficient)
ProgramGenesMotifsEnhancer: Transcription factor motifs enriched at top 300 gene's ABC linked enhancers (top 300 genes ranked by z-score coefficient)
ProgramGenesZScoreCoefficientGOTermsTop10: Top 10 gene ontology terms enriched for top 300 genes ranked by z-score coefficient
ProgramGenesMedianSpectraZScoreGOTermsTop10: Top 10 gene ontology terms enriched for top 300 genes ranked by raw weight (or called median spectra from cNMF)
median.spectra.zscore.df_k_*.dt_*/.txt
{enhancer, promoter}.topic.top.300.zscore.gene_motif.count.ttest.enrichment_motif.thr.{pval1e-4, pval1e-6, qval0.1}_k_*.dt_*.txt
clusterProfiler_GeneRankingType{zscore, raw, median_spectra, median_spectra_zscore}_EnrichmentType{ByWeightGSEA, GOEnrichment, GSEA, PosGenesGOEnrichment}.txt
sample.batch.correction.mtx.txt
summary.varianceExplained.df.txt
metrics.varianceExplained.df.txt
Perturb-seq outputs (The pipeline will generate these files only if "Perturb-seq" in config file is set to True:
*_MAST_DEtopics.txt
primerid Pr(>Chisq) coef ci.hi ci.lo fdr perturbation zlm.model.name fdr.across.ptb
topic_1 0.931958981499932 -0.00308004078521135 0.0570387818054911 -0.0631988633759138 0.997587143723284 A1BG batch.correction 0.980597200884276
topic_10 0.997587143723284 0.00216003169008006 0.0715983146968009 -0.0672782513166408 0.997587143723284 A1BG batch.correction 0.999373860636769
topic_11 0.996904491792907 -6.48785292078902e-06 0.0407059273499904 -0.040718903055832 0.997587143723284 A1BG batch.correction 0.99916248171282
topic_12 0.938744741036869 0.00810003101565848 0.0544634189392725 -0.0382633569079555 0.997587143723284 A1BG batch.correction 0.982593736237456
topic_13 0.0766707475946088 -0.0661311794734254 -0.00293861544807195 -0.129323743498779 0.33755308978221 A1BG batch.correction 0.390958645335121
topic_14 0.610995278682664 0.0246987961275237 0.091452821024267 -0.0420552287692196 0.945173197958634 A1BG batch.correction 0.867881082549678
Column description:
primerid: Program name
Pr(>Chisq): p-value
coef: Fold change (log2)
ci.hi: Confidence interval (upper bound)
ci.lo: Confidence interval (lower bound)
fdr: FDR across k programs for each gene
perturbation: Perturbed gene name
zlm.model.name: Name of the model fit to the data before statistical test. Supplied from workflow/scripts/MAST_model_formulas.txt (Default: ~condition + lane)
fdr.across.ptb: FDR across k programs and all genes
Outputs will be in config['analysisDir']/{cNMF_gene_selection}/{sampleName}/K*/threshold_*/program_prioritization_{GenomeWide, GWASWide}/{GWASTrait}/
*.program_prioritization.txt
Note that if "Perturb-seq" in the config file is set to "False", there will only be "ProgramGene" output. Regulator information requires perturbation significant{LinkedGenes, ProgramGene, Regulator}.formatted.df.txt