A Snakemake workflow to filter raw variants (snp and indels) and annotate vcf (variant call format) files of single samples (unrelated individuals) or cohort samples (related individuals) of paired-end sequencing data (WGS or WES) using GATK4, SnpSift, VEP, genmod and dbSNP. The vcf file can be optionally be prepared for ingestion into scout which involves the removal of multiallelic sites and scoring/ranking of variants. The pipeline can run on NVIDIA GPU's where nvidia clara parabricks software is available for significant speedups in analysis times. This pipeline is designed be used after human_genomics_pipeline and before the data is ingested into scout for clinical interpretation. However, this pipeline also stands on it's own, as a vcf annotation pipeline. This pipeline has been developed with human genetic data in mind, however we designed it to be species agnostic. Genetic data from other species can be analysed by setting a species-specific reference genome and variant databases in the configuration file (but not all situations have been tested).
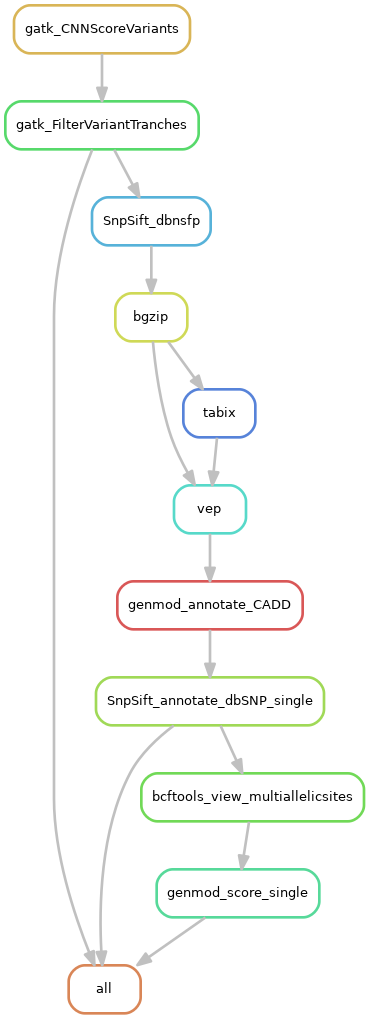
- Filter variants (gatk cnnscoreVariants and gatk FilterVariantTranches)
- Annotate variants with known information (with dbNSFP, vep, CADD, dbSNP databases)
- Prepare for scout (remove multiallelic sites, scoring/ranking of variants) (optional)
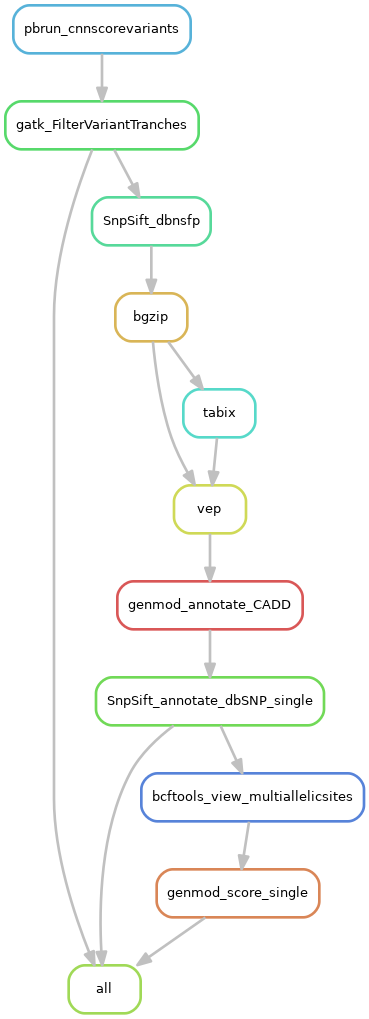
- Filter variants (parabricks CNNScoreVariants and gatk FilterVariantTranches)
- Equivalent to gatk cnnscoreVariants
- Annotate variants with known information (with dbNSFP, vep, CADD, dbSNP databases)
- Prepare for scout (remove multiallelic sites, scoring/ranking of variants) (optional)
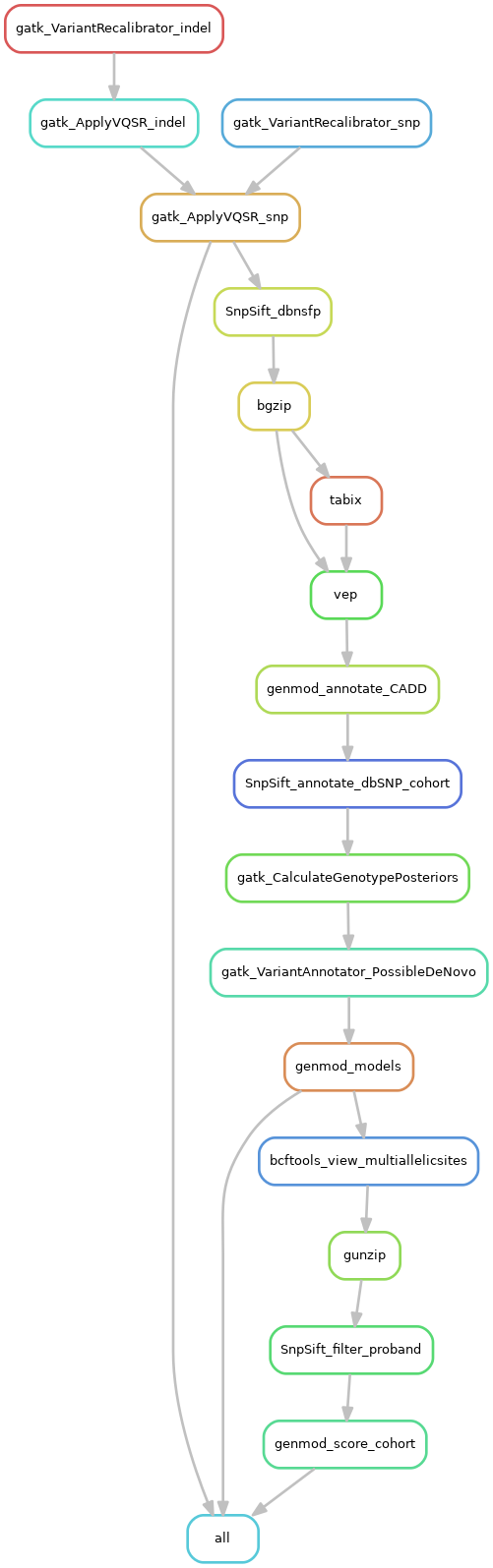
- Filter variants (gatk VariantRecalibrator and gatk ApplyVQSR)
- Annotate variants with known information (with dbNSFP, vep, CADD, dbSNP databases)
- Annotate variants with other information (genotype posterior probabilities, mark denovo variants, patterns of inheritance)
- Prepare for scout (remove multiallelic sites, filter for variants found in the proband, scoring/ranking of variants) (optional)
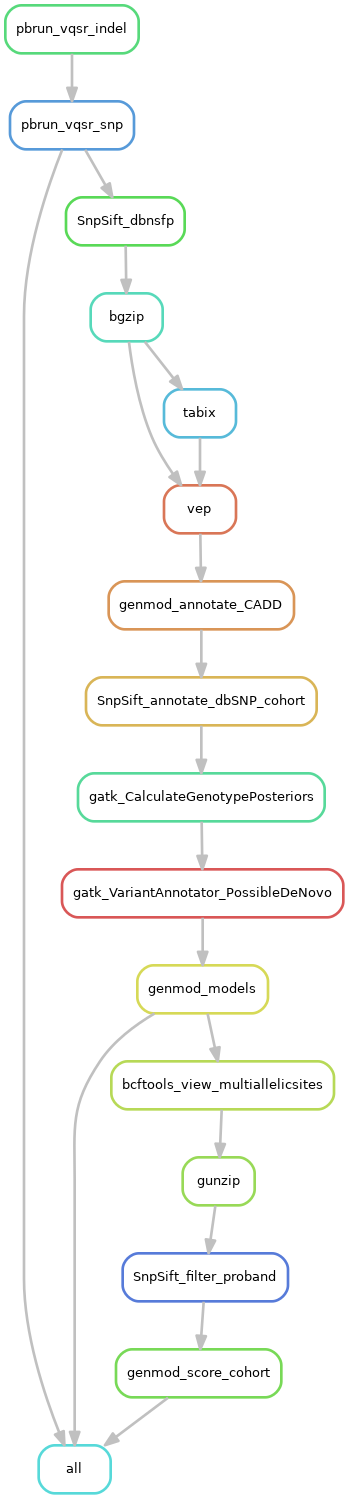
- Filter variants (pbrun vqsr)
- Equivalent to gatk VariantRecalibrator and gatk ApplyVQSR
- Annotate variants with known information (with dbNSFP, vep, CADD, dbSNP databases)
- Annotate variants with other information (genotype posterior probabilities, mark denovo variants, patterns of inheritance)
- Prepare for scout (remove multiallelic sites, filter for variants found in the proband, scoring/ranking of variants) (optional)
Single samples:
results/filtered/sample1_filtered.vcfresults/annotated/sample1_filtered_annotated.vcfresults/readyforscout/sample1_filtered_annotated_readyforscout.vcf.gz
Cohort samples:
results/filtered/sample1_filtered.vcfresults/annotated/sample1_filtered_annotated.vcfresults/readyforscout/sample1_filtered_annotated_readyforscout.vcf
- Prerequisite hardware: NVIDIA GPUs (for GPU accelerated runs)
- Prerequisite software: NVIDIA CLARA parabricks and dependencies (for GPU accelerated runs), Git (tested with version 2.7.4), Mamba (tested with version 0.4.4) with Conda (tested with version 4.8.2), gsutil (tested with version 4.52), gunzip (tested with version 1.6), R (tested with version 3.2.2)
The provided test dataset can be used to test running this pipeline on a new machine, or test pipeline developments/releases.
See the docs for a walkthrough guide for running vcf_annotation_pipeline on:
- Raise issues in the issues page
- Create feature requests in the issues page
- Start a discussion in the discussion page
- Contribute your code! Create your own branch from the development branch and create a pull request to the development branch once the code is on point!
Contributions and feedback are always welcome! 😊