Pytorch 1.4.0 code for:
Deep Active Learning for Joint Classification & Segmentation with Weak Annotator (https://arxiv.org/abs/2010.04889)
Citation:
@InProceedings{belharbi2020DeepAlJoinClSegWeakAnn,
title = {Deep Active Learning for Joint Classification \& Segmentation with Weak Annotator},
author = {Belharbi, S. and Ben Ayed, I. and McCaffrey, L. and Granger, E.},
booktitle = {Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision},
year = {2021}
}
Issues:
Please create a github issue.
Content:
Method highlights:
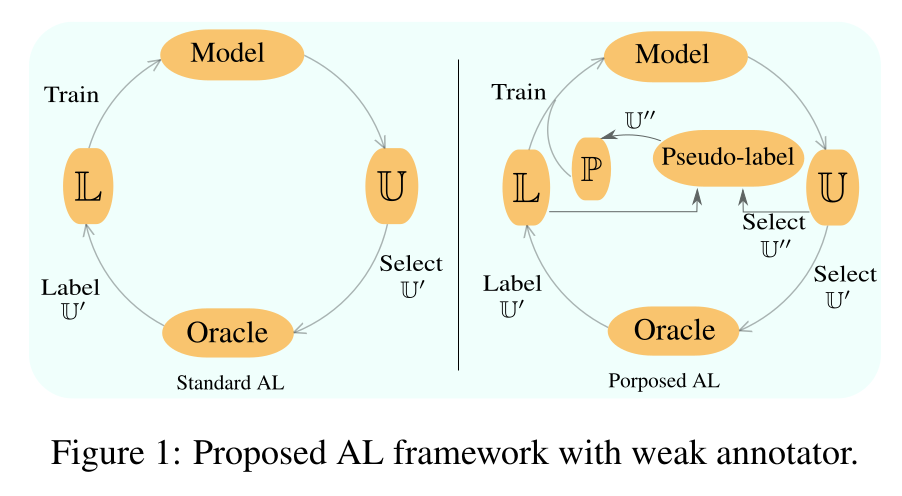
- Method:
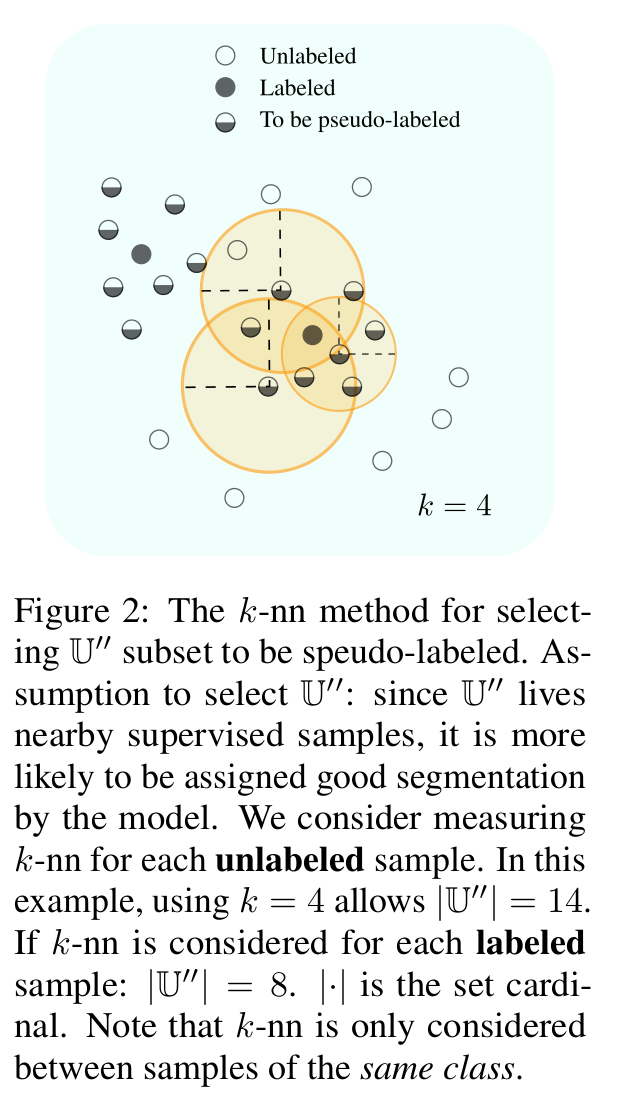
- Knn:
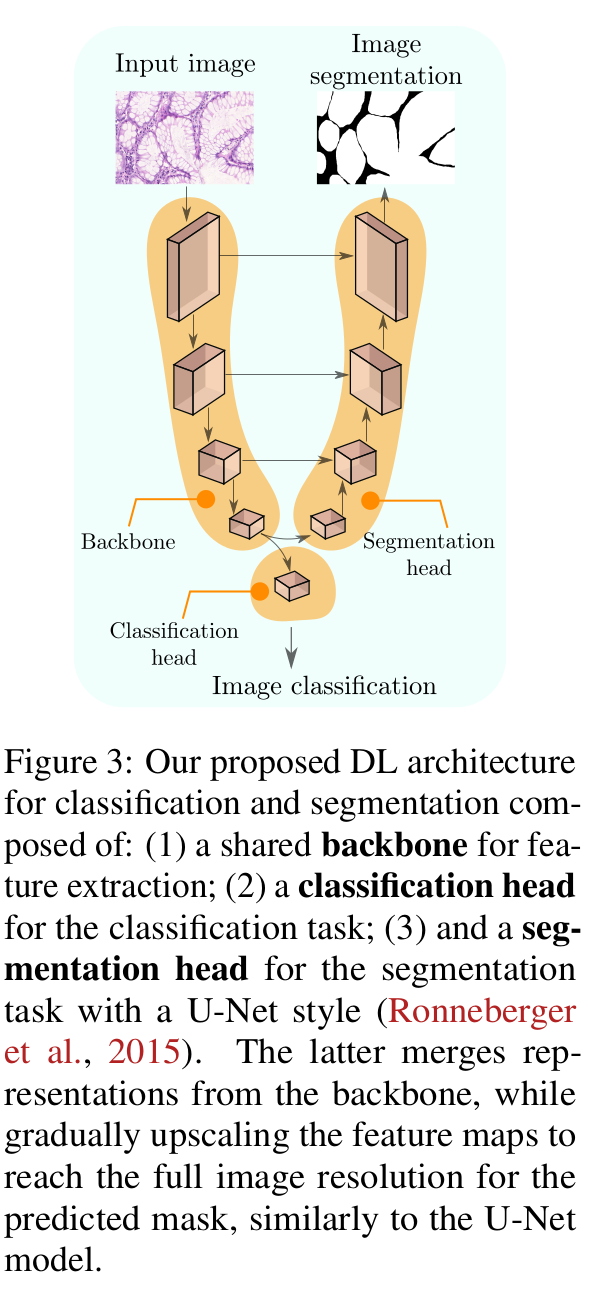
- Architecture:
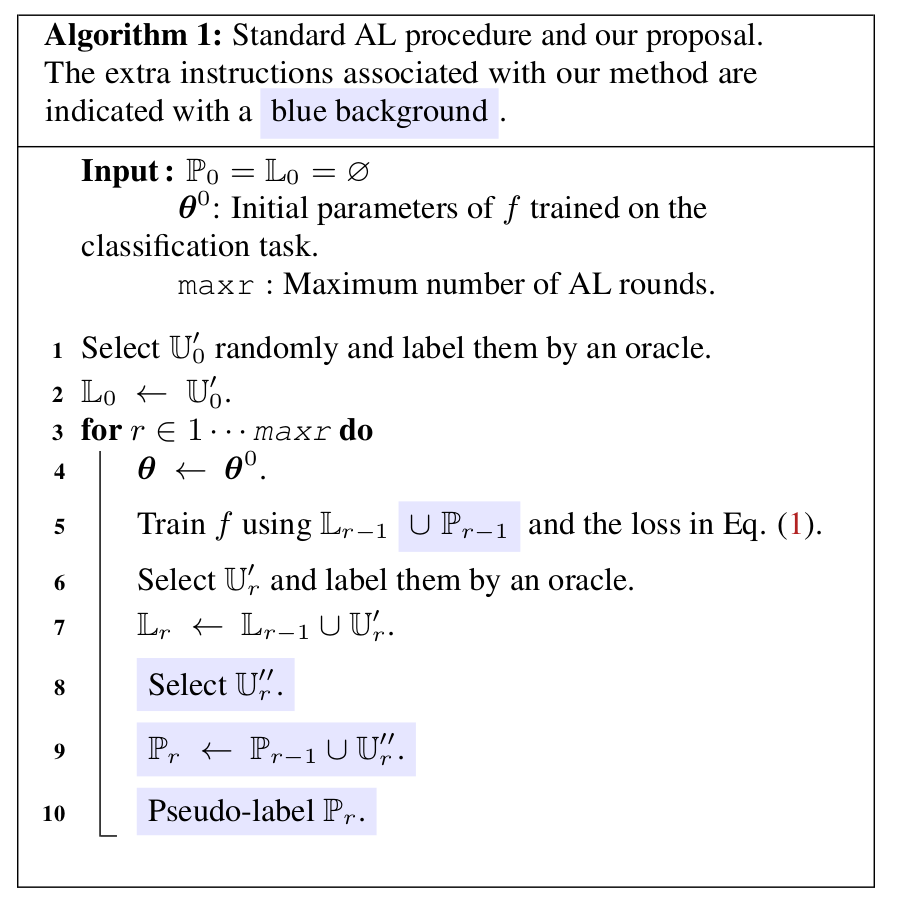
- Algorithm:
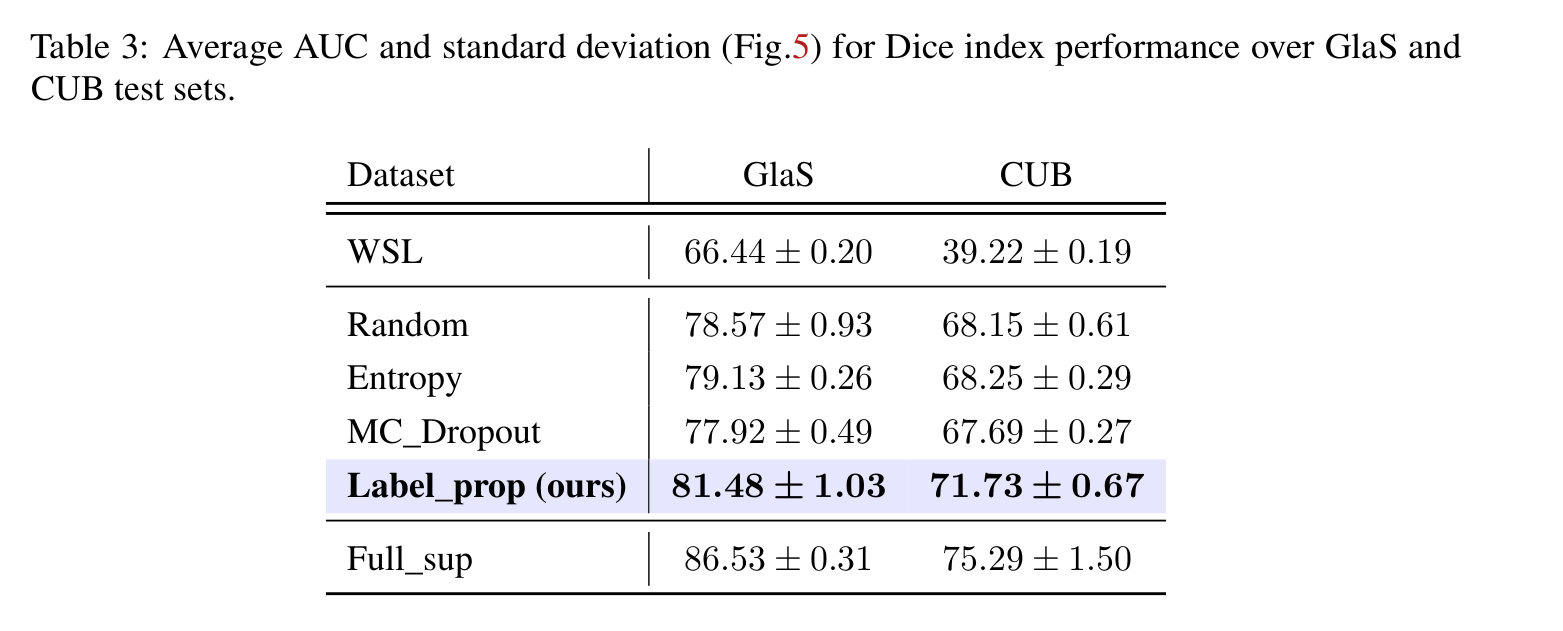
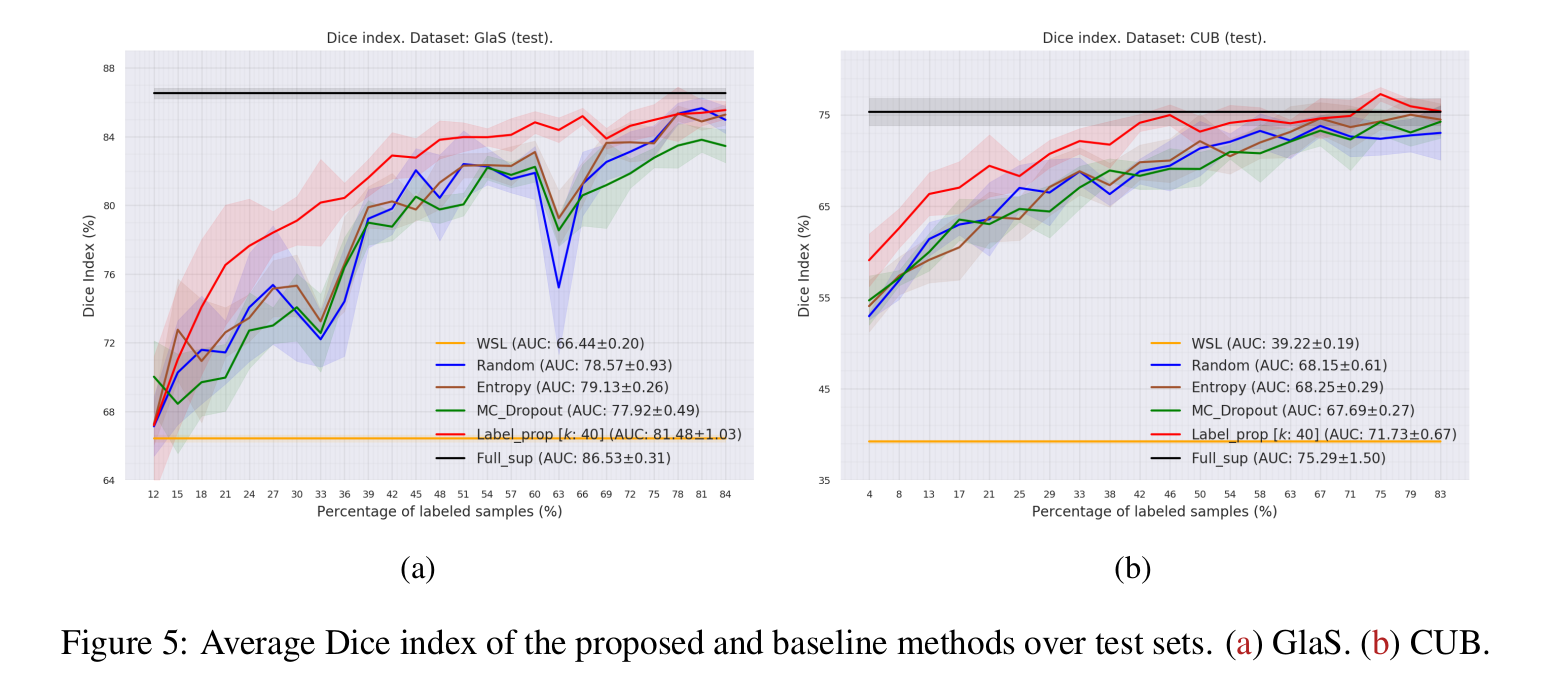
- Perfromance:
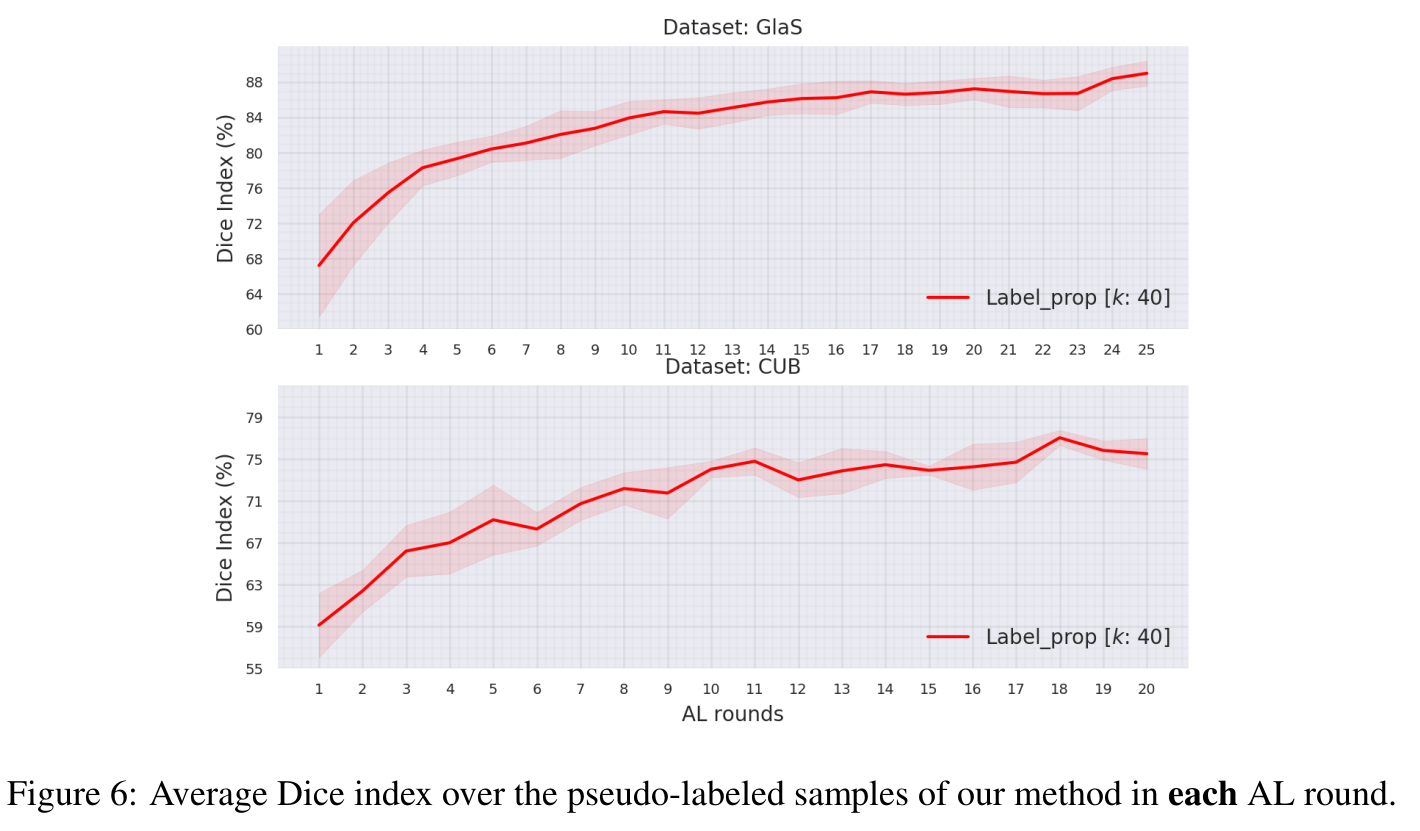
- Pseudo-labeling performance:
Download datasets:
- GlaS: ./download-glas-dataset.sh.
- Caltech-UCSD Birds-200-2011: ./download-caltech-ucsd-birds-200-2011-dataset.sh
You find the splits in ./folds. The code that generated the splits is ./create_folds.py.
Requirements:
We use Pytorch 1.4.0 and Python 3.7.6. For installation, see ./dependencies for a way on how to install the requirements within a virtual environment.
How to run the code step-by-step:
-
Generate the CSV files using:
python create_folds.py
-
Generate yaml files:
python yaml-gen.py
This will generate a yaml file per dataset. Each yaml provides only the backbone of the hyper-parmeters. We will update them in the next step according to each dataset/active learning method.
-
Generate the bash files to run the active learning experiments.
python gen_configs.py
This will generate all the experiments for all active learning methods for all the datasets (GLAS, CUB). For instance, if you want to run the method
MC_DropoutoverGLASdataset on the gpu0, use:./run-glas-MC_Dropout.sh 0
The file
run-glas-MC-Dropout.shcontains all the experiments for this method/datasets. It is something like this:#!/usr/bin/env bash # ============================================================================== cudaid=$1 # Start MYSEED: 0 time python main.py --yaml glas.yaml --MYSEED 0 --optn0__name_optimizer sgd \ --batch_size 20 --valid_batch_size 1 --optn0__lr 0.1 --optn0__weight_decay \ 0.0001 --optn0__momentum 0.9 --max_epochs 1000 --optn0__name_lr_scheduler \ mystep --optn0__lr_scheduler True --optn0__step_size 100 --optn0__gamma 0.9 \ --optn0__min_lr 1e-07 --optn0__t_max 50 --optn0__beta1 0.9 --optn0__beta2 \ 0.999 --optn0__eps_adam 1e-08 --optn0__amsgrad False --name_model \ hybrid_model --backbone resnet18 --backbone_dropout 0.0 --path_pre_trained \ ./pretrained/resnet18-glas.pt --pre_trained True --alpha 0.6 --kmax 0.1 \ --kmin 0.1 --dropout 0.1 --dropoutnetssl 0.5 --modalities 5 --crop_size 416 \ --ratio_scale_patch 0.9 --up_scale_small_dim_to 432 --padding_ratio 0.01 \ --pad_eval True --cudaid $cudaid --dataset glas --p_init_samples \ 11.940298507462687 --p_samples 2.985074626865672 --max_al_its 25 --task \ SEGMENTATION --split 0 --fold 0 --subtask Classification_Segmentation \ --nbr_bins_histc 256 --min_histc 0.0 --max_histc 1.0 --sigma_histc 100000.0 \ --resize_h_to_opt_mask 128 --resize_mask_opt_mask True --smooth_img True \ --smooth_img_ksz 3 --smooth_img_sigma 0.9 --enhance_color True \ --enhance_color_fact 3.0 --backbone_dropout 0.2 --mcdropout_t 50 \ --debug_subfolder paper_label_prop/glas/MC_Dropout --knn 40 \ --freeze_classifier True --segloss_l BinCrossEntropySegmLoss --segloss_pl \ BinCrossEntropySegmLoss --scale_cl 1.0 --scale_seg 1.0 --scale_seg_u 0.1 \ --scale_seg_u_sch ConstantWeight --scale_seg_u_end 1e-09 --scale_seg_u_sigma \ 150.0 --scale_seg_u_p_abstention 0.0 --weight_pseudo_loss False --protect_pl \ True --estimate_pseg_thres True --estimate_seg_thres True --seg_threshold 0.5 \ --base_width 24 --leak 64 --seg_elbon False --loss HybridLoss --al_type \ MC_Dropout --al_it 0 --exp_id 10_03_2020_13_58_33_376747__3089229 # ============================================================================== time python main.py --yaml glas.yaml --MYSEED 0 --optn0__name_optimizer sgd \ --batch_size 20 --valid_batch_size 1 --optn0__lr 0.1 --optn0__weight_decay \ 0.0001 --optn0__momentum 0.9 --max_epochs 1000 --optn0__name_lr_scheduler \ mystep --optn0__lr_scheduler True --optn0__step_size 100 --optn0__gamma 0.9 \ --optn0__min_lr 1e-07 --optn0__t_max 50 --optn0__beta1 0.9 --optn0__beta2 \ 0.999 --optn0__eps_adam 1e-08 --optn0__amsgrad False --name_model \ hybrid_model --backbone resnet18 --backbone_dropout 0.0 --path_pre_trained \ ./pretrained/resnet18-glas.pt --pre_trained True --alpha 0.6 --kmax 0.1 \ --kmin 0.1 --dropout 0.1 --dropoutnetssl 0.5 --modalities 5 --crop_size 416 \ --ratio_scale_patch 0.9 --up_scale_small_dim_to 432 --padding_ratio 0.01 \ --pad_eval True --cudaid $cudaid --dataset glas --p_init_samples \ 11.940298507462687 --p_samples 2.985074626865672 --max_al_its 25 --task \ SEGMENTATION --split 0 --fold 0 --subtask Classification_Segmentation \ --nbr_bins_histc 256 --min_histc 0.0 --max_histc 1.0 --sigma_histc 100000.0 \ --resize_h_to_opt_mask 128 --resize_mask_opt_mask True --smooth_img True \ --smooth_img_ksz 3 --smooth_img_sigma 0.9 --enhance_color True \ --enhance_color_fact 3.0 --backbone_dropout 0.2 --mcdropout_t 50 \ --debug_subfolder paper_label_prop/glas/MC_Dropout --knn 40 \ --freeze_classifier True --segloss_l BinCrossEntropySegmLoss --segloss_pl \ BinCrossEntropySegmLoss --scale_cl 1.0 --scale_seg 1.0 --scale_seg_u 0.1 \ --scale_seg_u_sch ConstantWeight --scale_seg_u_end 1e-09 --scale_seg_u_sigma \ 150.0 --scale_seg_u_p_abstention 0.0 --weight_pseudo_loss False --protect_pl \ True --estimate_pseg_thres True --estimate_seg_thres True --seg_threshold 0.5 \ --base_width 24 --leak 64 --seg_elbon False --loss HybridLoss --al_type \ MC_Dropout --al_it 1 --exp_id 10_03_2020_13_58_33_376747__3089229 # ============================================================================== ... and so on.
See all the keys that you can override using the command line in
parseit .get_yaml_args(). Seeconstants.pyfor the different active learning methods and other constants. -
First, run
WSLmethod using seed0. Then, store the model's weights in./pretrained/resnet18-glas.ptforGLASdataset, and./pretrained/resnet18-cub.ptforCUBdataset. The model's weight will be stored by the code in the experiment folder asbest_model.pt. Copy and rename that file to the pretrained folder. Then, you can proceed training other active learning methods. For convenience, we provide the pre-trained models. They are also accessible on google drive. If you are unable to access to the google drive folder, you can create an issue to find a different way to share the models. -
To plot all the curves of ALL methods, you can use,
python plot_active_learning_curves.py
Figures and other files will be stored in the folder
./paper. -
All experiments are done on split 0, fold 0 for all datasets.
-
Used seeds: 0, 1, 2, 3, 4.
-
For comparison, test Dice index at each round for each seed per method /dataset are stored in a pickled file
./paper/results-per-method-each-run -dataset-{}.pklin a dictionary with the key structure (for GlaS example):In [1]: import pickle as pkl In [20]: with open("results-per-method-each-run-dataset-glas.pkl", "rb") as fin: ...: stuff = pkl.load(fin) In [22]: stuff.keys() Out[22]: dict_keys(['WSL', 'Random', 'Entropy', 'MC_Dropout ', 'Label_prop ', 'Full_sup']) In [23]: stuff['Random'].keys() Out[23]: dict_keys(['test']) In [25]: stuff['Random']['test'].keys() Out[25]: dict_keys(['dice_idx']) In [30]: type(stuff['Random']['test']['dice_idx']) Out[30]: numpy.ndarray In [33]: stuff['Random']['test']['dice_idx'].shape Out[33]: (5, 25) # 5 runs ordered according to the seeds mentioned above. # each run has 25 active learning rounds. In [34]: stuff['Random']['test']['dice_idx'] Out[34]: array([[64.29491024, 68.26534145, 68.65601286, 73.98197889, 74.09444943, 76.94103431, 71.02902677, 74.06351458, 72.56103337, 78.99458144, 79.30639431, 82.01419845, 76.71336837, 84.38693479, 82.37608485, 82.45970614, 82.74210036, 70.06073229, 80.62327955, 81.25951763, 83.21664881, 85.40988602, 84.97116379, 85.60305871, 85.8245454 ], [68.31420053, 66.07314749, 68.52430042, 68.37349785, 69.61945057, 72.38897134, 69.81996067, 70.94271801, 72.44806387, 76.77195571, 77.22773626, 81.96324389, 81.54838331, 82.73607336, 82.25827597, 80.07024139, 79.83664479, 74.32793241, 83.06158803, 83.11748274, 83.19173351, 83.69847603, 85.22795111, 85.6280417 , 85.46123117], [66.71858971, 74.63980228, 70.05757164, 72.07348552, 76.8091803 , 74.19304218, 74.75387901, 71.29095811, 76.03948873, 80.75353745, 80.39951026, 82.95400567, 78.27928425, 78.79928647, 82.69563619, 81.45481266, 80.41814812, 72.53054075, 77.89946325, 81.37026601, 84.9570284 , 82.88242236, 84.4128149 , 86.50917932, 83.91877256], [69.44425181, 72.50236176, 74.71349549, 71.88765407, 71.65116169, 71.98040944, 76.57313634, 70.48917307, 71.09192843, 78.13675031, 80.39377347, 83.49617004, 82.48789147, 82.3112049 , 80.35638832, 81.90072447, 82.43326299, 80.88932212, 82.0582027 , 83.96295935, 81.2384364 , 83.12704161, 86.00610383, 85.92767775, 85.55987038], [66.9079571 , 69.86388833, 75.98011885, 70.86716615, 78.10507054, 81.32279277, 76.64835989, 74.21971507, 79.86538256, 81.44296072, 81.71520639, 79.7781498 , 83.15113798, 83.77281189, 83.63054134, 81.77533798, 84.02370453, 78.2903273 , 82.59848274, 82.92209167, 83.00199233, 83.72783348, 86.06250815, 84.60930221, 84.13211673]])
General notes:
- All the experiments, splits generation were achieved using seed 0. See ./create_folds.py
- All the results in the paper were obtained using one GPU.
Paths:
We hard-coded some paths (to the data location). For anonymization reasons, we replaced them with fictive paths. So, they won't work for you. A warning will be raised with an indication to the issue. Then, the code exits. It is something like this:
warnings.warn("You are accessing an anonymized part of the code. We are going to exit. Come here and fix this "
"according to your setup. Issue: absolute path to Caltech-UCSD-Birds-200-2011 dataset.")or
"Sorry, it seems we are enable to recognize the " \
"host. You seem to be new to this code. " \
"So, we recommend you add your baseurl on your own."