Official Pytorch Implementation for y-Aware Contrastive Learning (MICCAI 2021) [paper]
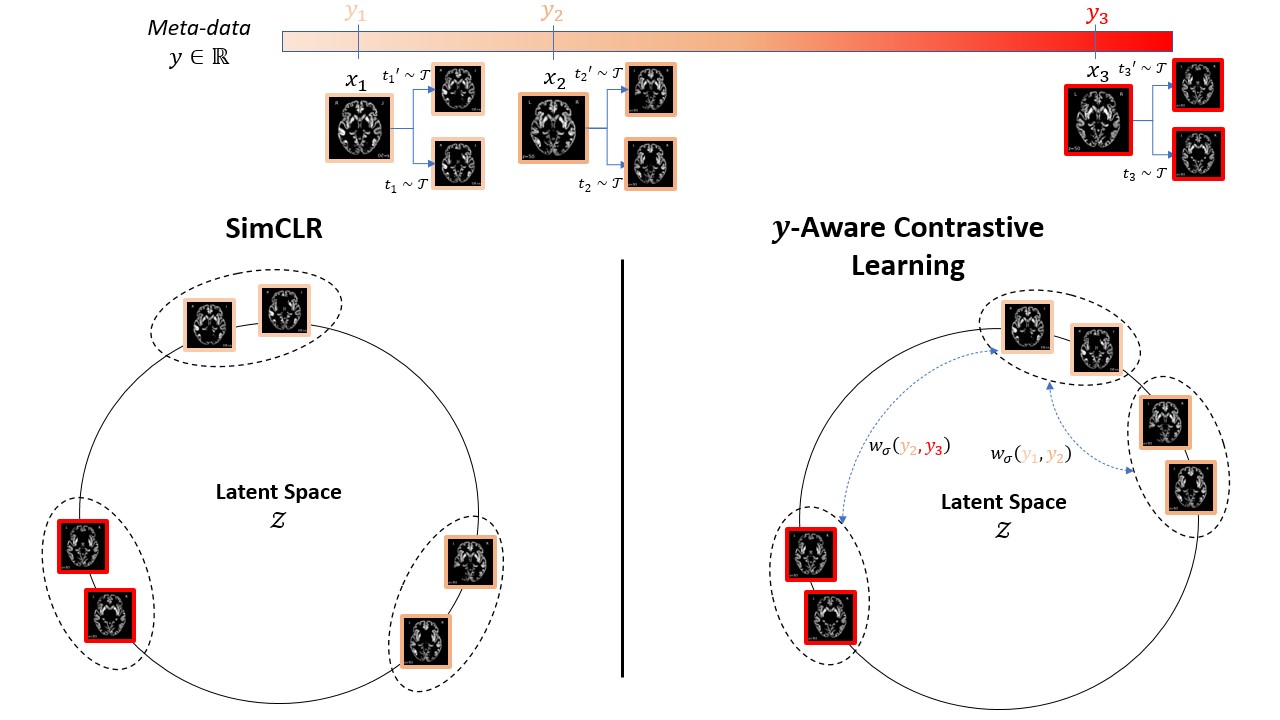
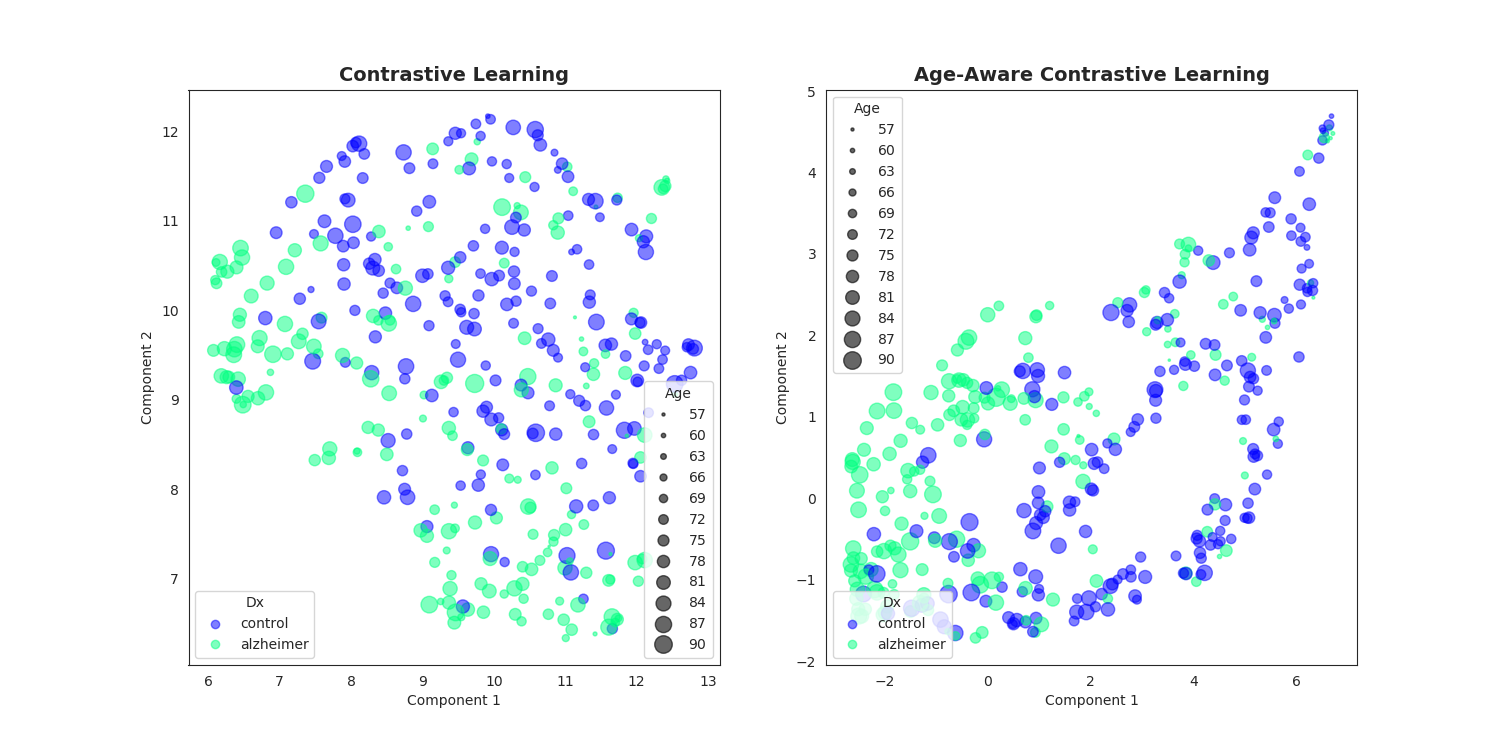
We propose an extension of the popular InfoNCE loss used in contrastive learning (SimCLR, MoCo, etc.) to the weakly supervised case where auxiliary information y is available for each image x (e.g subject's age or sex for medical images). We demonstrate a better data representation with our new loss, namely y-Aware InfoNCE.
- python >= 3.6
- pytorch >= 1.6
- numpy >= 1.17
- scikit-image=0.16.2
- pandas=0.25.2
In the paper, we aggregated 13 MRI datasets of healthy cohorts pre-processed with CAT12. You can find the complete list below.
| Source | # Subjects | # Sessions | Age | Sex (%F) | # Sites |
|---|---|---|---|---|---|
| HCP | 1113 | 1113 | 29 ± 4 | 45 | 1 |
| IXI | 559 | 559 | 48 ± 16 | 55 | 3 |
| CoRR | 1371 | 2897 | 26 ± 16 | 50 | 19 |
| NPC | 65 | 65 | 26 ± 4 | 55 | 1 |
| NAR | 303 | 323 | 22 ± 5 | 58 | 1 |
| RBP | 40 | 40 | 23 ± 5 | 52 | 1 |
| OASIS 3 | 597 | 1262 | 67 ± 9 | 62 | 3 |
| GSP | 1570 | 1639 | 21 ± 3 | 58 | 1 |
| ICBM | 622 | 977 | 30 ± 12 | 45 | 3 |
| ABIDE 1 | 567 | 567 | 17 ± 8 | 17 | 20 |
| ABIDE 2 | 559 | 580 | 15 ± 9 | 30 | 17 |
| Localizer | 82 | 82 | 25 ± 7 | 56 | 2 |
| MPI-Leipzig | 316 | 316 | 37 ± 19 | 40 | 2 |
| Total | 7764 | 10420 | 32 ± 19 | 50 | 74 |
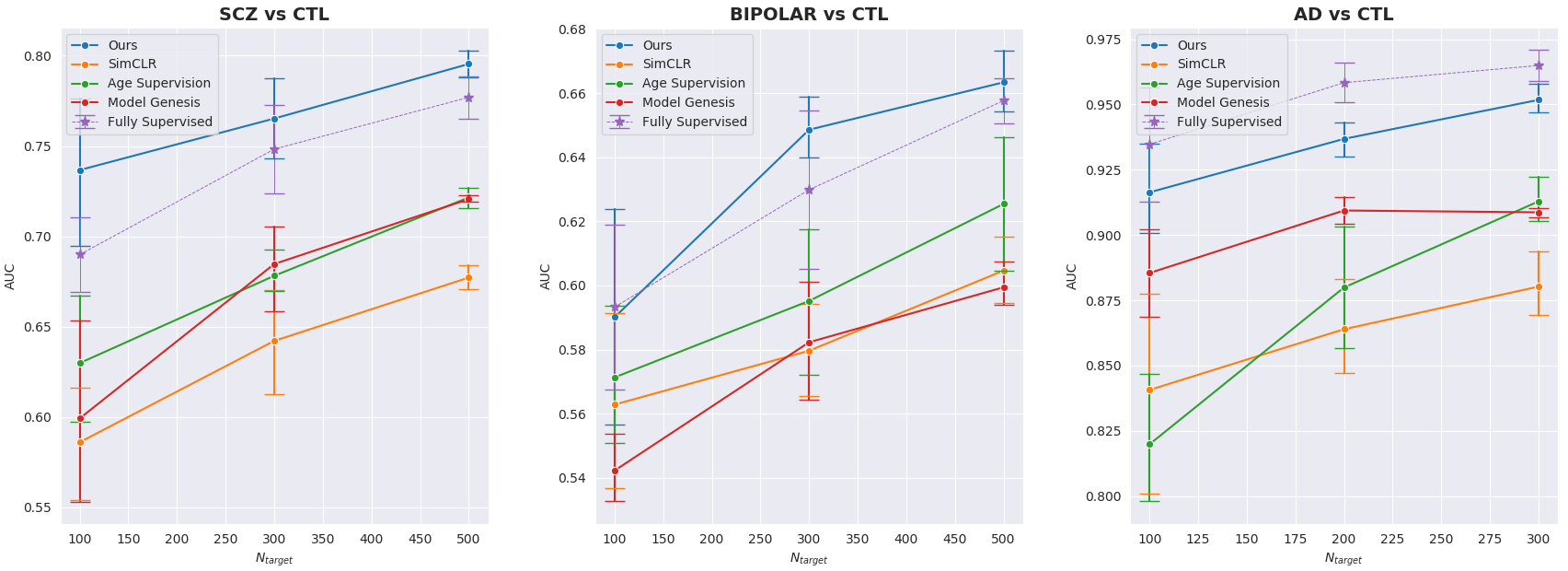
Originally, we have evaluated our approach on 3 classification target tasks with 2 public datasets (detailed below) and 1 private one (BIOBD). We also pre-processed the T1-MRI scan with CAT12 toolbox and all the images passed a visual Quality Check (QC).
| Source | # Subjects | Diagnosis | Age | Sex (%F) | # Sites |
|---|---|---|---|---|---|
| ADNI-GO | 387 | Alzheimer |
75 ± 8 |
52 |
57 |
| SCHIZCONNECT-VIP | 605 | Schizophrenia |
34 ± 12 |
27 |
4 |
First, you can clone this repository with:
$ git clone https://github.com/Duplums/yAwareContrastiveLearning.git
$ cd yAwareContrastiveLearningYou can download our DenseNet121 model pre-trained on BHB-10K here.
We have used only random cutout during pre-training and we used the hyperparameters defined by default in config.py.
Then you can directly run the main script with your configuration in config.py including:
- the paths to your training/validation data
- the proxy label you want to use during training along with the hyperparameter sigma
- the network (critic) including a base encoder and a projection head which is here a simple MLP(2)
self.data_train = "/path/to/your/training/data.npy"
self.label_train = "/path/to/your/training/metadata.csv"
self.data_val = "/path/to/your/validation/data.npy"
self.label_val = "/path/to/your/validation/metadata.csv"
self.input_size = (C, H, W, D) # typically (1, 121, 145, 121) for sMRI
self.label_name = "age" # asserts "age" in metadata.csv columns
self.checkpoint_dir = "/path/to/your/saving/directory/"
self.model = "DenseNet"Once you have filled config.py with the correct paths, you can simply run the DenseNet model with:
$ python3 main.py --mode pretrainingIn order to fine-tune the model on your target task, do not forget to set the path to the downloaded file in config.py:
self.pretrained_path = "/path/to/DenseNet121_BHB-10K_yAwareContrastive.pth"Then you can define your own Pytorch Dataset in main.py:
dataset_train = Dataset(...)
dataset_val = Dataset(...)You can finally fine-tune your model with:
$ python3 main.py --mode finetuning