Official Code Repository for the paper "Graph Self-supervised Learning with Accurate Discrepancy Learning" (NeurIPS 2022): https://arxiv.org/abs/2202.02989
In this repository, we implement Discrepancy-based graph Self-supervised LeArning (D-SLA).
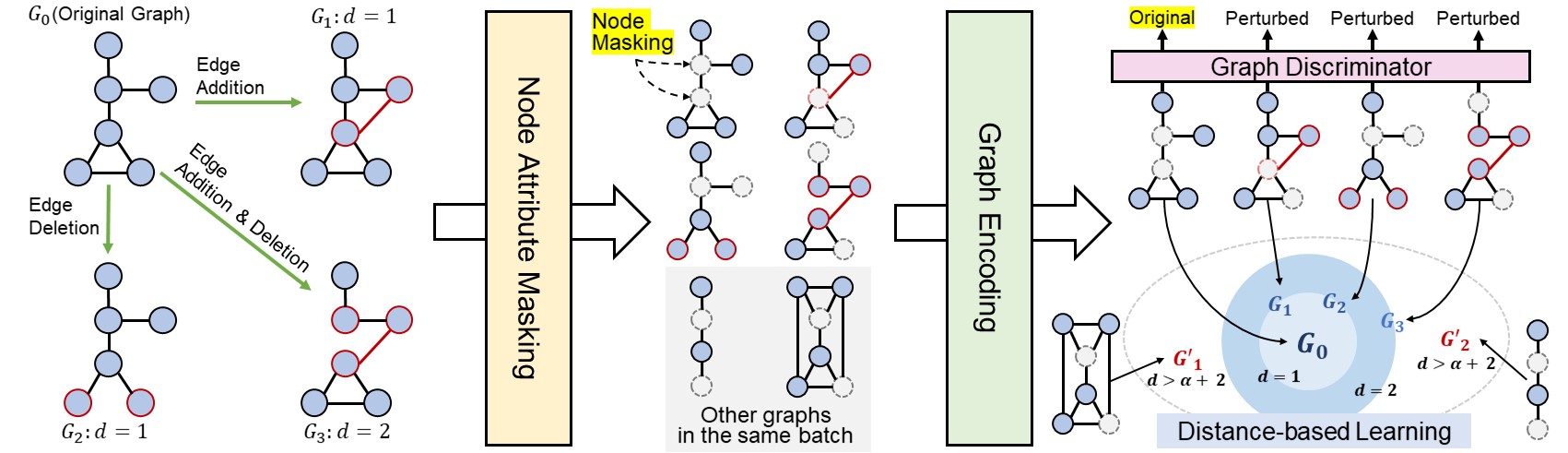
- We propose a novel graph self-supervised learning framework with a completely opposite objective from contrastive learning, which aims to learn to differentiate a graph and its perturbed ones using a discriminator, as even slight perturbations could lead to completely different properties for graphs.
- Utilizing the graph edit distance that is obtained for perturbed graphs at no cost, we propose a novel objective to preserve the exact amount of discrepancy between graphs in the representation space.
- We validate our D-SLA by pre-training and fine-tuning it on various benchmarks of chemical, biology, and social domains, on which it significantly outperforms baselines.
python 3.8
pytroch 1.7.0
torch-geometric 1.7.0
torch-cluster 1.5.9
torch-scatter 2.0.6
torch-sparse 0.6.9
torch-spline-conv 1.2.1
rdkit 2022.03.5
To download the molecular graph dataset, run commands below
wget http://snap.stanford.edu/gnn-pretrain/data/chem_dataset.zip
unzip chem_dataset.zip -d transferLearning_MoleculeNet_PPI/chem/To download the biological graph dataset, run commands below
wget http://snap.stanford.edu/gnn-pretrain/data/bio_dataset.zip
unzip bio_dataset.zip -d transferLearning_MoleculeNet_PPI/bio/cd transferLearning_MoleculeNet_PPI/chem
python pretrain_DSLA.py --edit_learn --add_strong_pert --margin_learn --margin {MARGIN} --lr {LR}For example,
python pretrain_DSLA.py --edit_learn --add_strong_pert --margin_learn --margin 5.0 --lr 0.001cd transferLearning_MoleculeNet_PPI/bio
python pretrain_DSLA.py --edit_learn --add_strong_pert --margin_learn --margin 10.0 --lr 0.001The pair of (DATASET_NAME, PERTURBATION_STRENGTH) is the one of (COLLAB, 0.001), (IMDB-BINARY, 0.01), (IMDB-MULTI, 0.01).
cd linkPrediction_TU
python pretrain_DSLA.py --dataset {DATASET_NAME} --edge_pert_strength {PERTURBATION_STRENGTH} --edit_learnFor example,
python pretrain_DSLA.py --dataset COLLAB --edge_pert_strength 0.001 --edit_learnThe downstream dataset is the one of [bbbp, clintox, muv, hiv, bace, sider, tox21, toxcast].
cd transferLearning_MoleculeNet_PPI/chem
python finetune.py --model_file {MODEL_FILE} --runseed {SEED} --dataset {DATASET_NAME} --lr 0.001For example,
python finetune.py --model_file ckpts/DSLA/100.pth --runseed 0 --dataset bbbp --lr 0.001cd transferLearning_MoleculeNet_PPI/bio
python finetune.py --model_file {MODEL_FILE} --runseed {SEED} --lr 0.001For example,
python finetune.py --model_file ckpts/DSLA/100.pth --runseed 0 --lr 0.001The dataset is the one of [COLLAB, IMDB-BINARY, IMDB-MULTI].
cd linkPrediction_TU
python finetune_edgepred.py --dataset {DATASET_NAME} --model_file {MODEL_FILE} --seed {SEED}For example,
python finetune_edgepred.py --dataset COLLAB --model_file ckpts/COLLAB/DSLA/100.pth --seed 0@article{kim2022dsla,
author = {Dongki Kim and
Jinheon Baek and
Sung Ju Hwang},
title = {Graph Self-supervised Learning with Accurate Discrepancy Learning},
journal = {arXiv:2202.02989},
year = {2022},
url = {https://arxiv.org/abs/2202.02989}
}