Qin Zhu, Kim Lab, University of Pennsylvania
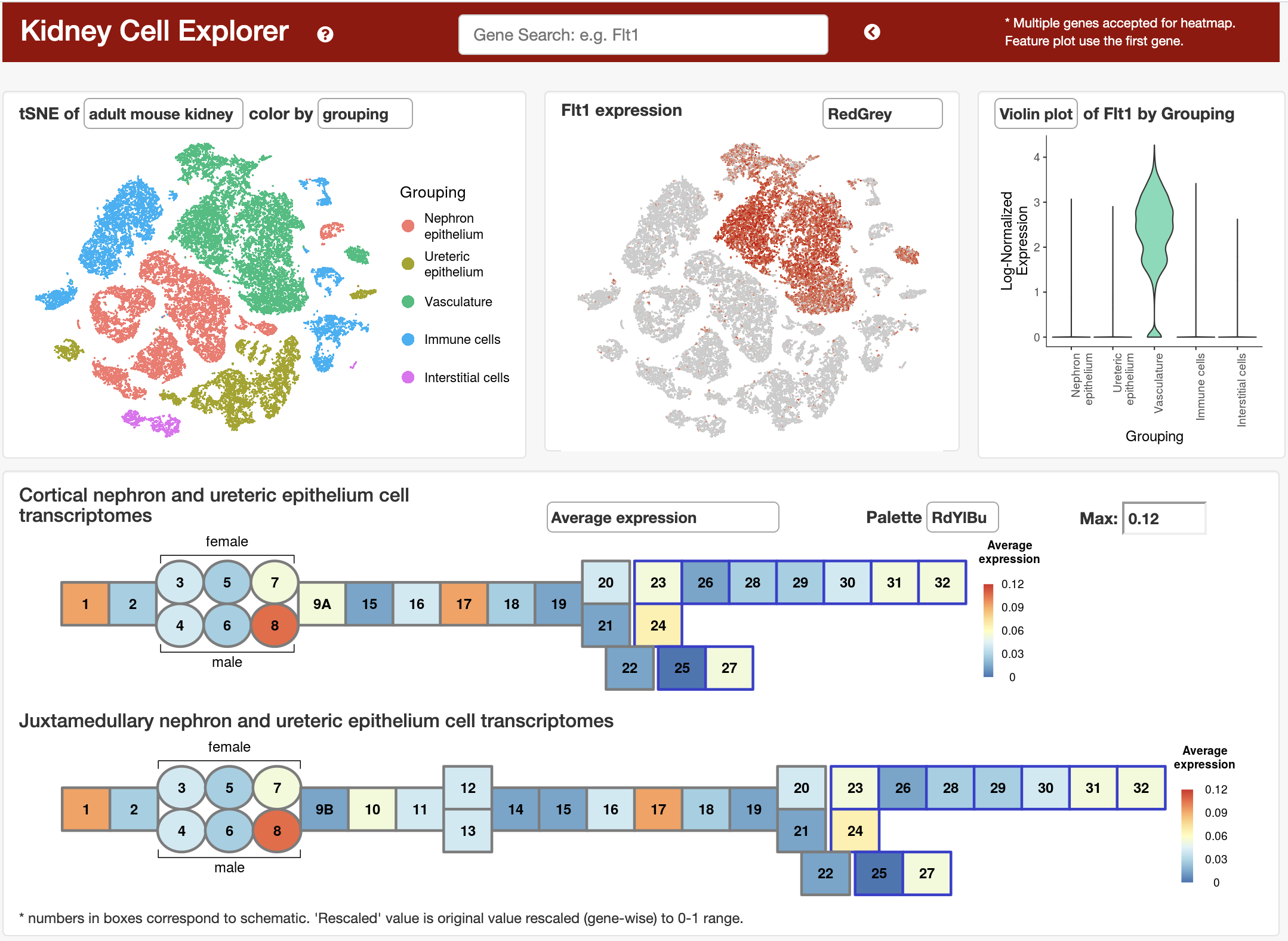
Kidney Cell Explorer (https://cello.shinyapps.io/kidneycellexplorer/) was developed using Shiny (Chang et al., 2018) and the R programming language. We utilized interactive visualization modules of the VisCello package (Packer et al., 2019, https://github.com/qinzhu/VisCello) and adapted them to allow exploration of kidney specific data. Key features of the website include interactive t-SNE plots, feature expression plots, metacell expression heatmap and detailed ontology annotation Expression heatmaps on the webpage show gene expression pattern across nephron and ureteric epithelium cells stratified by ontology based “metacells.”
https://cello.shinyapps.io/kidneycellexplorer/
Ransick, A., Lindström, N.O., Liu, J., Zhu, Q., Guo, J.J., Alvarado, G.F., Kim, A.D., Black, H.G., Kim, J. and McMahon, A.P., 2019. Single-Cell Profiling Reveals Sex, Lineage, and Regional Diversity in the Mouse Kidney. Developmental cell, 51(3), pp.399-413.