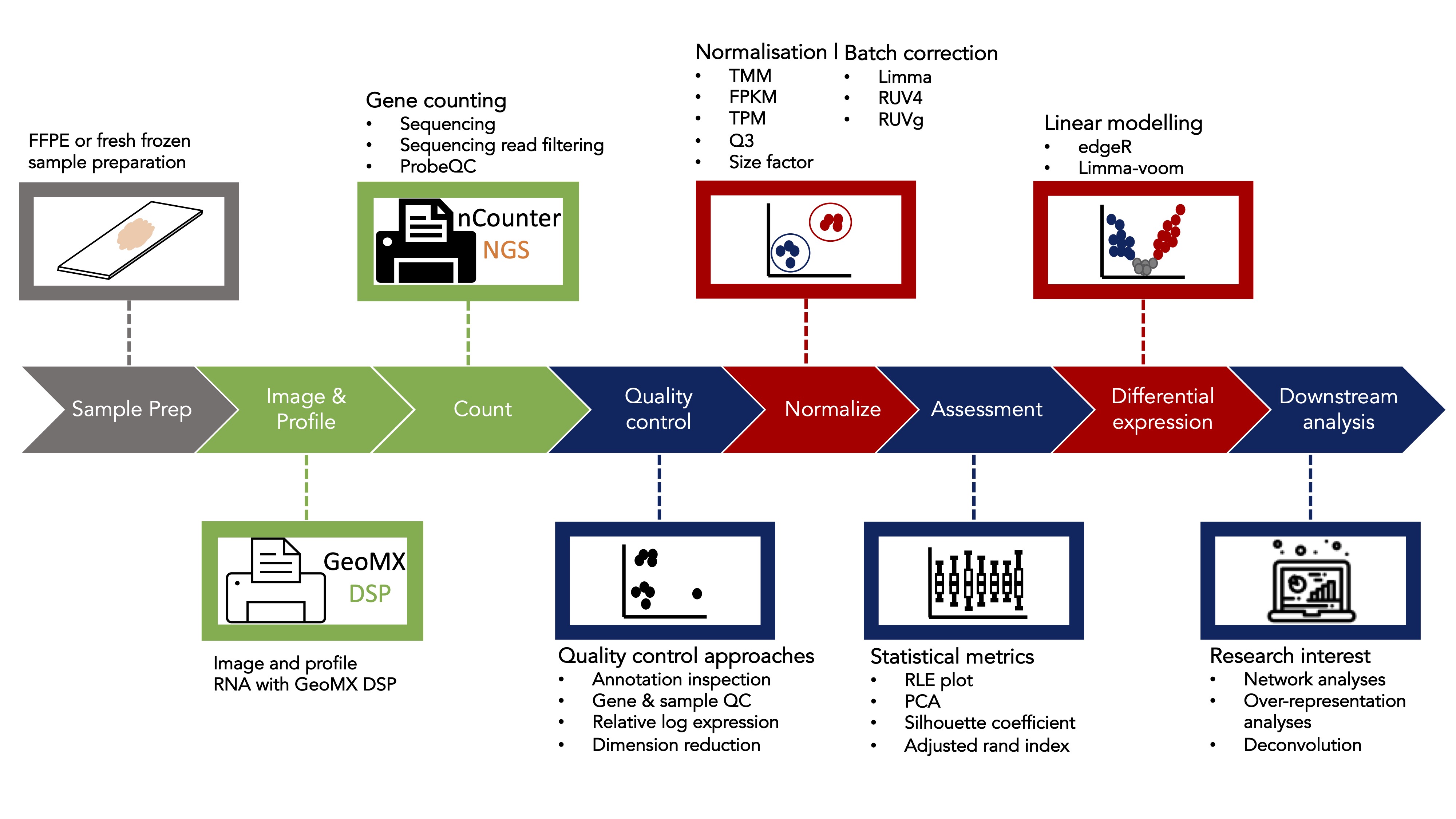
The standR package provides a series of functions to assist different stages of the analysis of spatial transcriptomics data generated from the Nanostring's GeoMX DSP platforms. Most functions in the package is based on the infrastructure SpatialExperiment, see detials here.
See full guide at HERE
The published GeoMX WTA data of diabetic kidney disease that we used in the vignette is available here.
Install the release version from Bioconductor
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("standR")
Install development version from GitHub
library(devtools)
devtools::install_github("DavisLaboratory/standR")
To cite standR, plese cite the following article: https://doi.org/10.1093/nar/gkad1026 Much appreciated!