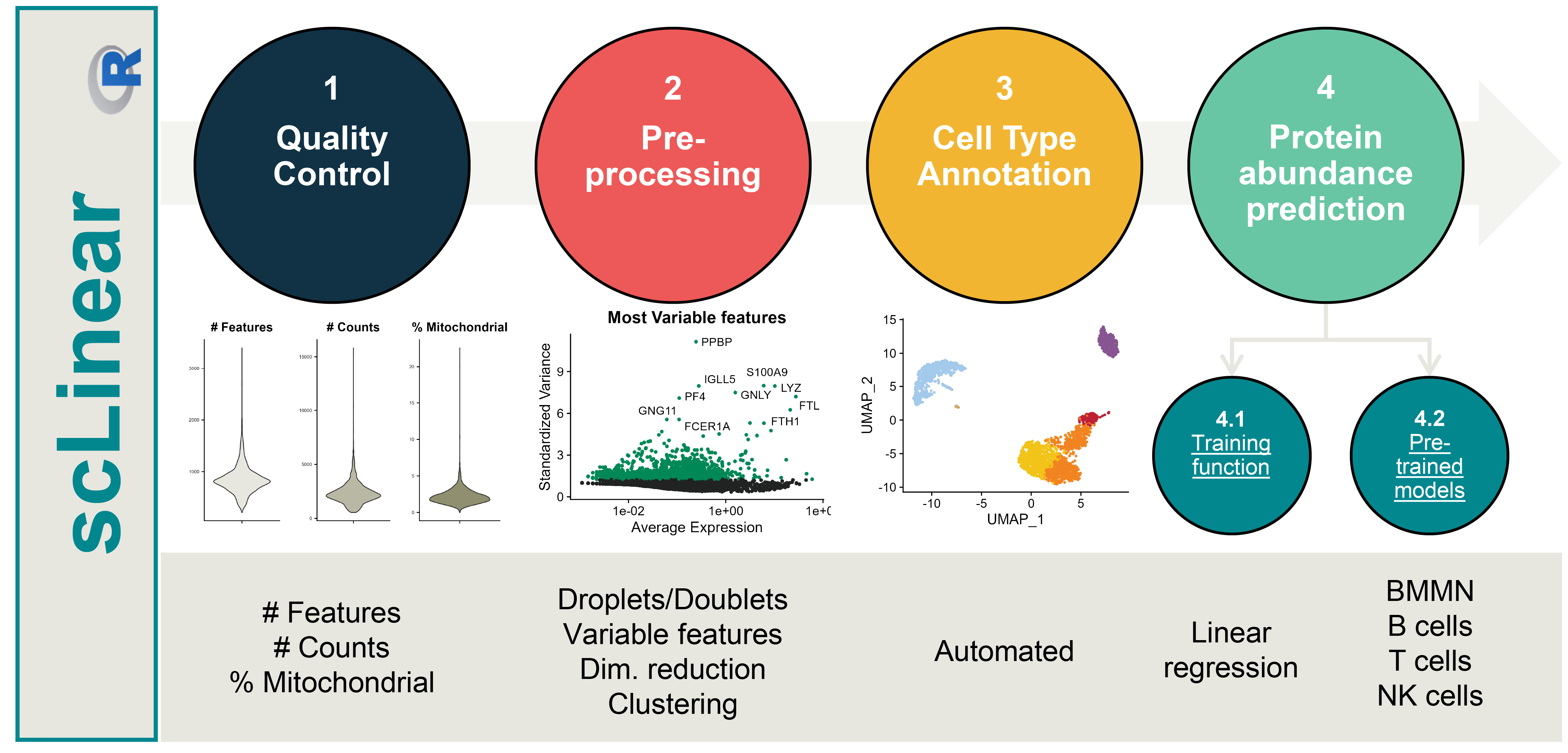
The goal of scLinear is to predict antibody derived tags (ADT) data from gene expression data in scRNA-seq data. It includes all the necessary pre-processing steps, comes equiped with pre-trained models and also allows the training of new models.
You can install the development version of scLinear using devtools. Because of multiple issues with python dependencies not correctly installed during package installation, I removed the automated installation, of the dependencies for now and added them to the installation guide (I will try to solve the issue in the future)
# Set up a python environment. I will use the renv package to create a new local environment. In case you want to use your default environment and r library, the first two commands can be scipped.
renv::init("./")
renv::use_python()
install.packages("devtools")
install.packages("reticulate")
# Get sure that the correct python environment is used by reticulate
reticulate::py_config()
#> python: /path/to/renv/python/virtualenvs/renv-python-3.10/bin/python
#> libpython: /usr/lib/python3.10/config-3.10-x86_64-linux-gnu/libpython3.10.so
#> pythonhome: /path/to/renv/python/virtualenvs/renv-python-3.10:/path/to/renv/python/virtualenvs/renv-python-3.10
#> version: 3.10.12 (main, Nov 20 2023, 15:14:05) [GCC 11.4.0]
#> numpy: [NOT FOUND]
#>
#> NOTE: Python version was forced by RETICULATE_PYTHON
# Install all python dependencies (The version specified for scikit-learn & numpy should be yoused, if you want to use the pretrained models)
py_modules_list <- c("numpy<1.26.0","joblib","scikit-learn==1.2.0","anndata","warnings","scanpy")
for (m in py_modules_list){
if(!reticulate::py_module_available(m)){reticulate::py_install(m)}
}
if(!reticulate::py_module_available("pytorch_lightning")){reticulate::py_install("pytorch_lightning", pip = TRUE)} # install with pip
# Test if all python dependencies are available
py_modules_list_available <- c("numpy","joblib","sklearn","anndata","warnings","torch","scanpy","os","scipy","typing", "pytorch_lightning")
py_modules_list_not_available <- c()
for (m in py_modules_list_available){
if(!reticulate::py_module_available(m)){py_modules_list_not_available <- c(py_modules_list_not_available,m)}
}
if(is.null(py_modules_list_not_available)){ "All python modules are available"}else{
print(paste0("The following python modules are not available: ", paste0(py_modules_list_not_available, collapse = ", ")))
print("Try installing them with: reticulate::py_install(missing module name)")
}
#> [1] "All python modules are available"
# if the installation of scLinear fails, try restarting the R session, to reload the reticulate environment
# Install scLinear
if (!require("devtools", quietly = TRUE)){install.packages("devtools")}
devtools::install_github("DanHanh/scLinear")The PBMC data can be downloaded from the 10X Genomics website (https://support.10xgenomics.com/single-cell-gene-expression/datasets/3.0.0/pbmc_10k_protein_v3). Then the following code can be used to generate a Seurat object.
library(scLinear)
set.seed(42)
# File: "Feature / cell matrix (filtered)"
# Download the cell matrix file into the local directory and untar it
dir.create("local", showWarnings = FALSE)
url <- "https://cf.10xgenomics.com/samples/cell-exp/3.0.0/pbmc_10k_protein_v3/pbmc_10k_protein_v3_filtered_feature_bc_matrix.tar.gz"
destfile <-"local/pbmc_10k_protein_v3_filtered_feature_bc_matrix.tar.gz"
download.file(url, destfile)
untar(destfile, exdir = "local")
# Create a Seurat object from the data
data_dir <- "local/filtered_feature_bc_matrix"
pbmc10k.data <- Seurat::Read10X(data.dir = data_dir)
rownames(x = pbmc10k.data[["Antibody Capture"]]) <- gsub(pattern = "_[control_]*TotalSeqB", replacement = "", x = rownames(x = pbmc10k.data[["Antibody Capture"]]))
pbmc10k <- Seurat::CreateSeuratObject(counts = pbmc10k.data[["Gene Expression"]], min.cells = 1, min.features = 1)
pbmc10k[["ADT"]] <- Seurat::CreateAssayObject(pbmc10k.data[["Antibody Capture"]][, colnames(x = pbmc10k)])
Seurat::DefaultAssay(pbmc10k) <- "RNA"
saveRDS(pbmc10k, "./local/pbmc10k.rds")You may run scLinear directly providing the Seurat object as input. List of optional parameters:
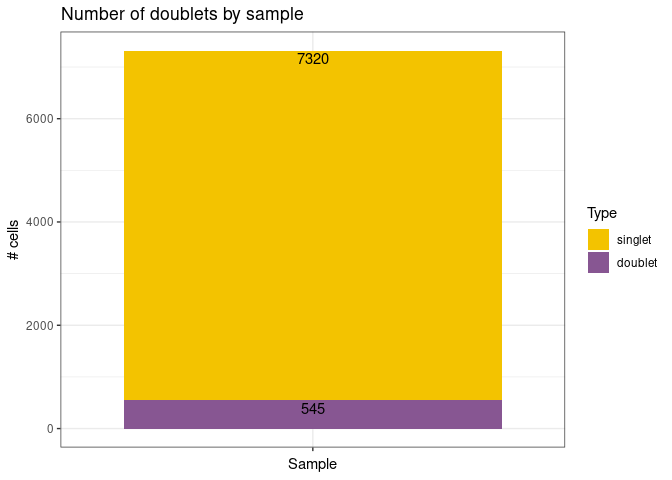
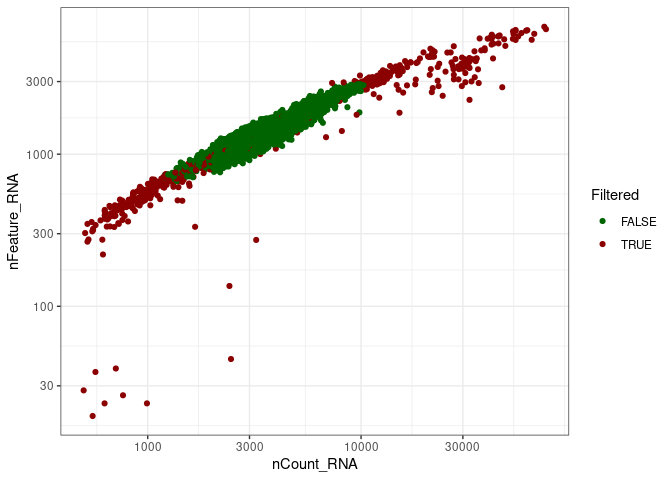
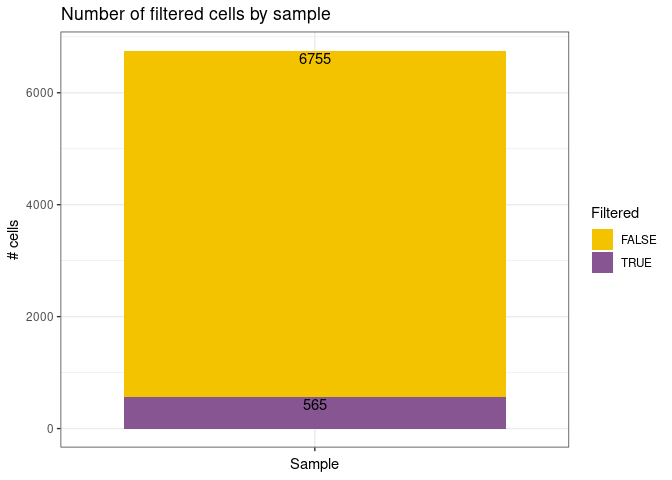
remove_doubletsRemoval of doublets. TRUE (default) or FALSE.low_qc_cell_removalRemoval of low quality cells. TRUE (default) or FALSE.anno_levelLevel of annotation. 1, 2, 3 or 4. See https://github.com/JiaLiVUMC/scMRMA for more details.samplesNULL (default).integrate_dataTRUE or FALSE (default).remove_empty_dropletsRemoval of empty droplets. TRUE or FALSE (default).lower= 100 (default).FDR= 0.01 (default).annotation_selfClusterTRUE or FALSE (default).resolution= 0.8 (default).seed= 42 (default).return_plotsTRUE or FALSE (default).modelAvailable models “all” (default), “bcell”, “tcell” and “nkcell”.assay_name= “RNA” (default).print_plotsTRUE or FALSE (default).
The scLinear function uses the counts slot from the RNA assay to predict the ADT assay. The functions performs the default preprocessing steps and returns a Seurat object with the added “predicted_ADT” assay
pbmc10k <- readRDS("./local/pbmc10k.rds")
pbmc10k_adt_predicted <- scLinear(pbmc10k)
#> [1] "Start remove doublets"
#> [1] "Start low quality cell removal"
#> [1] "Start clustering data"
#> [1] "Number of used dimensions for clustering: 25"
#> [1] "Start cell type annotation"
#> Pre-defined cell type database panglaodb will be used.
#> Multi Resolution Annotation Started.
#> Level 1 annotation started.
#> Level 2 annotation started.
#> Level 3 annotation started.
#> Level 4 annotation started.
#> Uniform Resolution Annotation Started.scLinear calls different sub-workflows which can also be called independently.
pbmc10k <- readRDS("./local/pbmc10k.rds")
pbmc10k <- prepare_data(pbmc10k,
integrate_data = FALSE,
annotation_selfCluster = TRUE,
remove_empty_droplets = FALSE)
#> [1] "Start remove doublets"#> [1] "Start low quality cell removal"
#> [1] "Start clustering data"
#> [1] "Number of used dimensions for clustering: 25"
#> [1] "Start cell type annotation"
#> Pre-defined cell type database panglaodb will be used.
#> Multi Resolution Annotation Started.
#> Level 1 annotation started.
#> Level 2 annotation started.
#> Level 3 annotation started.
#> Level 4 annotation started.
#> Uniform Resolution Annotation Started.
saveRDS(pbmc10k ,"./local/pbmc10k_prepared.rds")User may manually load pre-trained models (available models: all, bcell,
tcell, nkcell). If a pretrained model is used it is advided to use the
raw data slot from the RNA assay, and normalization = TRUE, to ensure
that the input data is normalized the same way as for the training data.
adt_predict() is then used to predict the ADT values with
parameters:
* pipe Pretrained model. * gexp Gene expression matrix. * slot
Seurat slot to use. “counts” (default) * normalize TRUE (default) or
FALSE. An example can be found below:
pbmc10k <- readRDS("./local/pbmc10k_prepared.rds")
pipe <- create_adt_predictor()
pipe <- load_pretrained_model(pipe, model = "all")
pbmc10k@assays["predicted_ADT"] <- adt_predict(pipe = pipe,
gexp = pbmc10k@assays[["RNA"]],
normalize = TRUE)
saveRDS(pbmc10k ,"./local/pbmc10k_predicted.rds")To train a new model the following commands need to be used.
create_adt_predictor() to initialize predictor.
fit_predictor() with parameters: * pipe predictor initialized
above.
* gexp_train gene expression matrix of training set (i.e. RNA assay
from Seurat object).
* adt_train ADT matrix of training set.
* normalize_gex Gene expression normalization. TRUE (default) or
FALSE. * normalize_adt ADT normalization. TRUE (default) or FALSE.
Subsequently, the evaluate_predictor command can be used (same
parameters with fit_predictor) to return the RMSE, Pearson and
Spearman of the training process. An example of this process can be
found below.
pbmc10k <- readRDS("./local/pbmc10k_prepared.rds")
## Create a training and a test set
set.seed(42)
indx <- sample(1:length(colnames(pbmc10k)), size = length(colnames(pbmc10k)), replace = FALSE)
pbmc10k_train <- pbmc10k[,indx[1:5000]]
pbmc10k_test <- pbmc10k[,indx[5001:length(colnames(pbmc10k))]]
## create predictor
pipe <- create_adt_predictor()
## train predictor
pipe <- fit_predictor(pipe = pipe,
gexp_train = pbmc10k_train@assays[["RNA"]],
adt_train = pbmc10k_train@assays[["ADT"]],
normalize_gex = TRUE,
normalize_adt = TRUE)
## save the trained model
save_trained_model(pipe = pipe, file = "./local/trained_model.joblib")
#> NULL
# load the trained model
pipe <- create_adt_predictor()
pipe <- load_pretrained_model(pipe, file = "./local/trained_model.joblib")
## evaluate predictor
eval_res <- evaluate_predictor(pipe = pipe,
gexp_test = pbmc10k_test@assays[["RNA"]],
adt_test = pbmc10k_test@assays[["ADT"]],
normalize_gex = TRUE,
normalize_adt = TRUE)
#> RMSE: 0.35243118253148364
#> Pearson correlation: 0.9411908520410573
#> Spearman correlation: 0.8725401159661871
print(eval_res)
#> RMSE Pearson Spearman
#> 1 0.3524312 0.9411909 0.8725401
## add the predicted adt assay
pbmc10k_test[["predicted_ADT"]] <- adt_predict(pipe = pipe,
gexp = pbmc10k_test@assays[["RNA"]],
normalize = TRUE)Daniel Hanhart et al., “ScLinear predicts protein abundance at single-cell resolution”
DOI: https://doi.org/10.1038/s42003-024-05958-4
For any request or question you may contact: * Daniel Hanhart daniel.hanhart@unibe.ch * Panagiotis Chouvardas panagiotis.chouvardas@unibe.ch