This repository contains the algorithm and the codes developed to predict a TILs-score in breast cancer histhopathology H&E slides. The training and the test data is provided by the TiGER challenge team. Our model is ranked 5th in the final leaderboard of the TiGER challenge with a C-index (concordance index) of 0.6034. The winning algorithm resulted in a C-index of 0.6388.
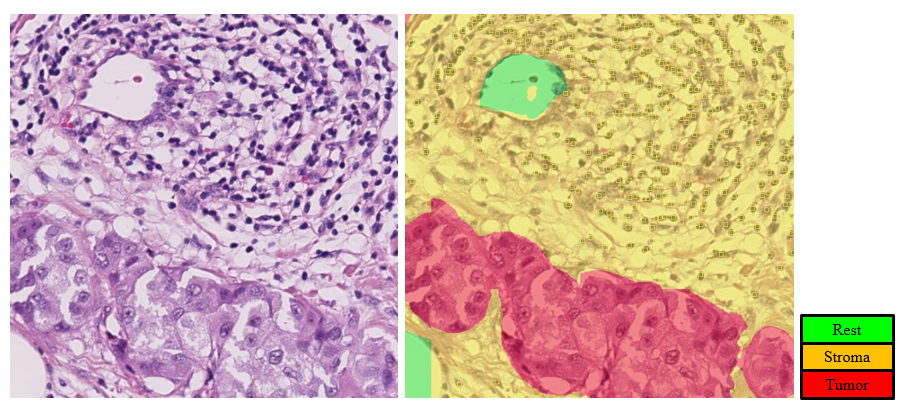
To design a TILs-score, we first need to segment the tissue into stromal and tumor regions. We then need to identify the TILs on the tissue regions. One example is shown in the figure below:
Using the following equation we can obtain the TILs-score. The TILs-score is calculated as the density of the TILs area within areas of tumor-associated stroma; the density ranges from 0 to 100:
To scan a whole slide image, we extract patches of size 256 with a stride of 256 in a for loop along the x-axis and another for loop along the y-axis. For each of the extracted patches, we predict a segmentation mask and detect the TILs.
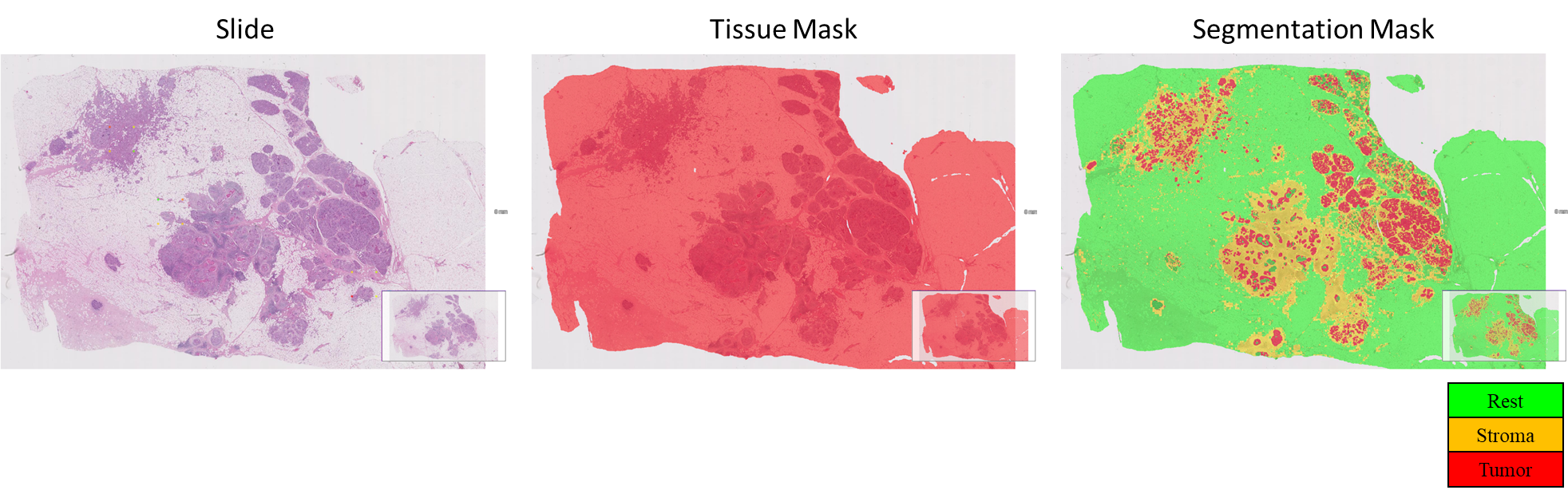
Here, as an example, one slide image along with the tissue mask is shown. Tissue mask identifies the regions of the tissue, everything else is the slide background. On the tissue region the algorithm identifies the stromal and tumor regions along with the TILs. As a result, the TILs-score can be obtained:
This code is made by Arian Arab as part of the Division of Imaging, Diagnostics and Software Reliability of the United States Food and Drug Administration. This code is based on the code developed by the TIGER challenge organizers.