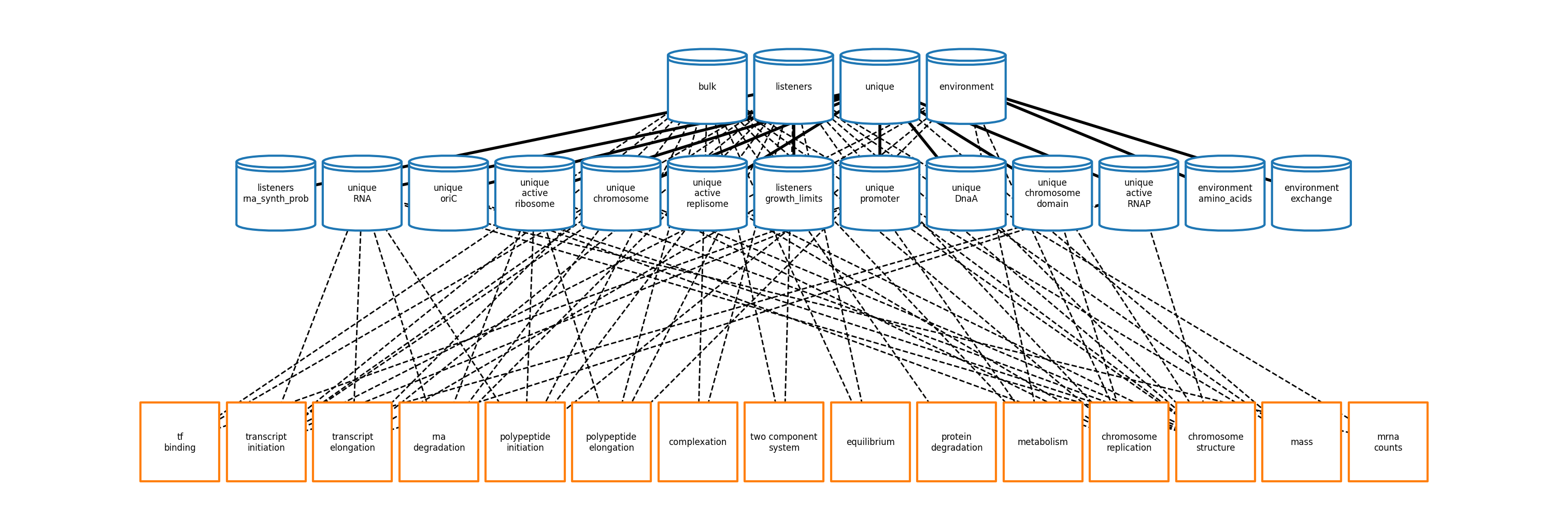
Vivarium E. coli (vEcoli) is a port of the Covert Lab's E. coli Whole Cell Model (wcEcoli) to the Vivarium framework. Its main benefits over the original model are:
- Modular processes: easily add/remove processes that interact with existing or new simulation state
- Unified configuration: all configuration happens through JSON files, making it easy to run simulations/analyses with different options
- Parquet output: simulation output is in a widely-supported columnar file format that enables fast, larger-than-RAM analytics with DuckDB
As in wcEcoli, raw experimental data is first processed by the parameter calculator or ParCa to calculate model parameters (e.g. transcription probabilities). These parameters are used to configure processes that are linked together into a complete simulation.
Note: The following instructions assume a Linux or MacOS system. Windows users can attempt to follow the same instructions after setting up Windows Subsystem for Linux.
Note: The instructions to set up the model on Sherlock are different and documented under the "Sherlock" sub-heading in the "Workflows" documentation page.
pyenv lets you install and switch between multiple Python releases and multiple "virtual environments", each with its own pip packages. Using pyenv, create a virtual environment and install Python 3.11.3. For a tutorial on how to install pyenv and other dependencies, follow the instructions here. Then, run the following command in your terminal:
pyenv virtualenv 3.11.3 viv-ecoli && pyenv local viv-ecoli
Update pip, setuptools and wheel to avoid issues with these:
pip install --upgrade pip setuptools wheel
Now, install numpy (check requirements.txt for the exact version):
pip install numpy==1.26.4
Then install the remaining requirements:
pip install -r requirements.txt
And build the Cython components:
make clean compile
Install nextflow following the instructions here.
After installing Java with SDKMAN!, close and reopen your terminal, then run
the following commands and compare their output to ensure that the Java compiler
and JVM were properly installed and the same version.
javac -version
java -version
After verifying your Java installation, you can proceed with installing Nextflow,
starting with curl -s https://get.nextflow.io | bash. If this fails, try
prepending export CAPSULE_LOG=verbose and re-run, checking for failed downloads.
If any downloads failed, re-run this command until it succeeds.
To test your installation, from the top-level of the cloned repository, invoke:
# Must set PYTHONPATH and OMP_NUM_THREADS for every new shell
export PYTHONPATH=.
export OMP_NUM_THREADS=1
python runscripts/workflow.py --config ecoli/composites/ecoli_configs/test_installation.json
This will run the following basic simulation workflow:
- Run the parameter calculator to generate simulation data.
- Run the simulation
for a single generation, saving output in
outfolder. - Analyze simulation output by creating a mass fraction plot.
Check out the user guide for a high-level tutorial of model development, details on key model components, and low-level API documentation.