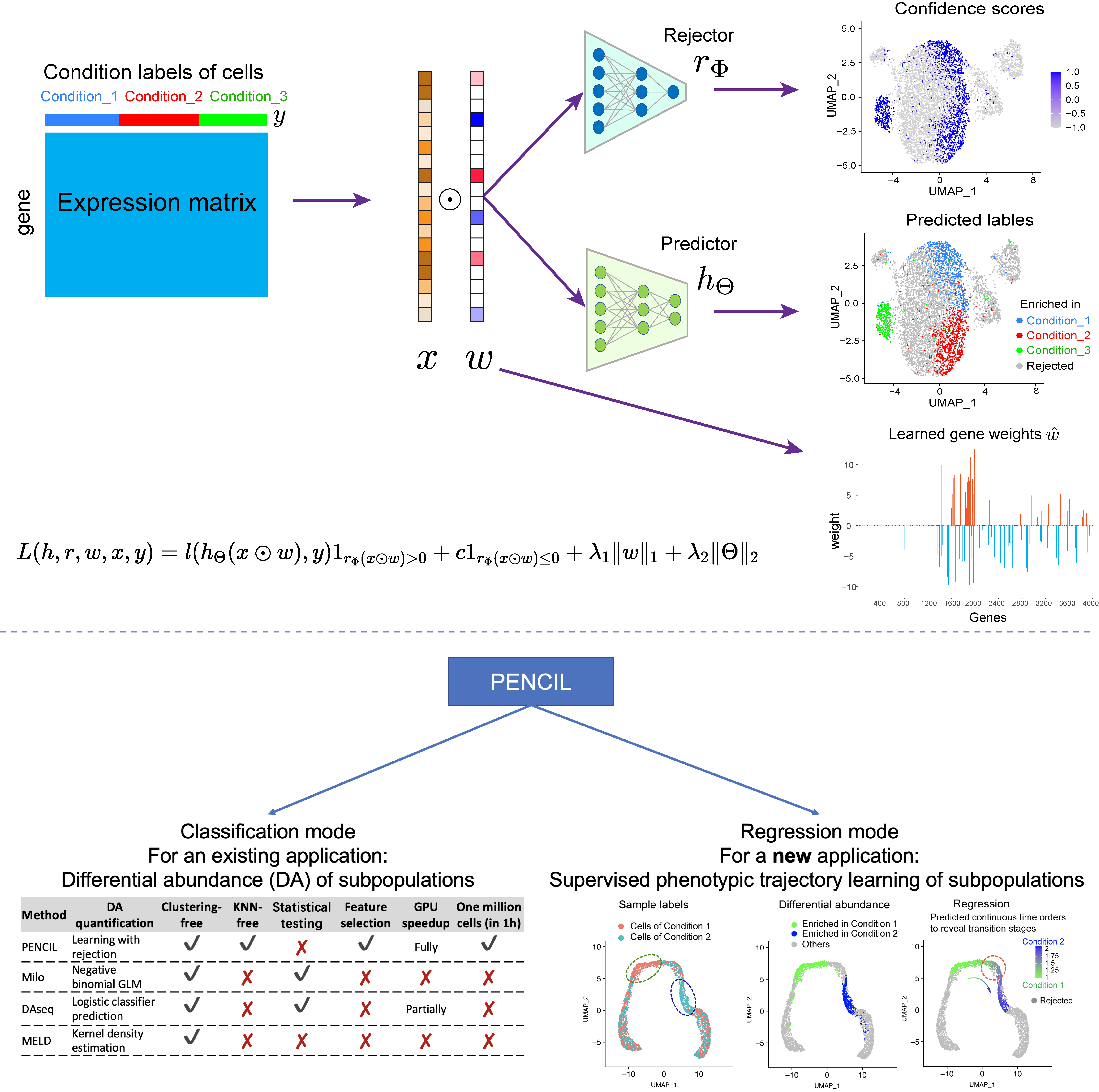
PENCIL is a supervised learning framework to identify high-confidence phenotype-associated subpopulations from single-cell data (PENCIL). PENCIL is flexible to take either categorical phenotypes or continuous variables as inputs by changing the prediction function. By embedding a feature selection function into this flexible framework, PENCIL is able to select informative features and identify phenotypic cell subpopulations simultaneously. The workflow of PENCIL is depicted in the following figure:
- Sep, 2022: PENCIL version 1.0.0 is launched.
PENCIL package requires only a standard computer with enough RAM to support the in-memory operations. If a GPU with enough VRAM is available, PENCIL can also be deployed on it to achieve computational acceleration.
The following runtimes are generated using a computer with 16GB RAM, 8 cores@3.2 GHz CPU, RTX3060 GPU (6GB VRAM) and 50 Mbps internet speed.
The developmental version of the package has been tested on the following systems:
- Windows
- MacOS
- Linux
PENCIL depends on the following Python packages:
numpy 1.20.3
pandas 1.3.4
torch 1.10.0
seaborn 0.11.2 p
umap-learn 0.5.2
mlflow 1.23.1
PENCIL is developed under Python(version >= 3.9). To build PENCIL, clone the repository:
git clone https://github.com/cliffren/PENCIL.git
cd PENCIL
Then install the PENCIL package by pip, and all requirements will be installed automatically.
pip install -e .
You can also install the dependency packages manually, especially for the GPU version of pytorch, which is automatically installed for the CPU version. The default installation should take approximately 1 minutes and 45 seconds on the computer for testing.
from pencil import *
# prepare data source
expression_data = np.random.rand(5000, 2000) # 5000 cells and 2000 genes.
phenotype_labels = np.random.randint(0, 3, 5000)
class_names = ['class_1', 'class_2', 'class_3']
# init a pencil model
model = Pencil(mode='multi-classification', select_genes=True, mlflow_record=True)
# run
with mlflow.start_run():
pred_labels, confidence = model.fit_transform(
expression_data, phenotype_labels,
class_names=class_names,
plot_show=True
)
gene_weights = model.gene_weights(plot=True)Using two examples with categorical or continuous phenotype labels, respectively, we demonstrate how to execute PENCIL.
If you are used to working with the R, start here:
If you also would like to use PENCIL in python, continue here:
The R tutorial or python tutorial would take about 5 minutes on the test computer using GPU (58 minutes using CPU only).
Please cite the following manuscript:
Supervised learning of high-confidence phenotypic subpopulations from single-cell data. Nature Machine Intelligence (2023). https://doi.org/10.1038/s42256-023-00656-y.
Tao Ren, Canping Chen, Alexey V. Danilov, Susan Liu, Xiangnan Guan, Shunyi Du, Xiwei Wu, Mara H. Sherman, Paul T. Spellman, Lisa M. Coussens, Andrew C. Adey, Gordon B. Mills, Ling-Yun Wu and Zheng Xia
PENCIL is licensed under the GNU General Public License v3.0.
PENCIL will be updated frequently with new features and improvements. If you have any questions, please submit them on the GitHub issues page or check the FAQ list.