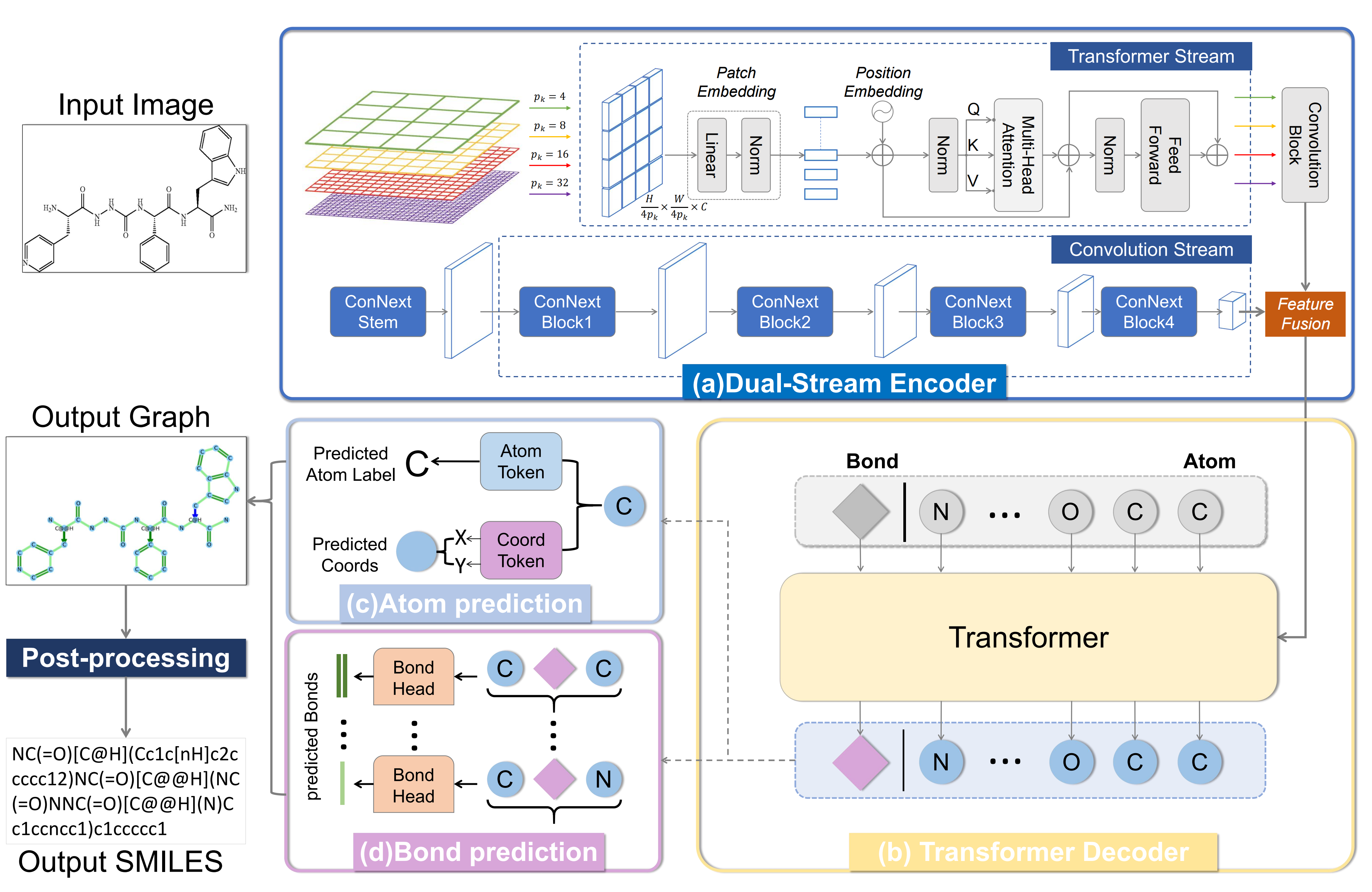
This is the offical code of following paper "MolNexTR: A Generalized Deep Learning Model for Molecular Image Recognition".
In this work, We propose MolNexTR, a novel graph generation model. The model follows the encoder-decoder architecture, takes three-channel molecular images as input, outputs molecular graph structure prediction, and can be easily converted to SMILES. We aim to enhance the robustness and generalization of the molecular structure recognition model by enhancing the feature extraction ability of the model and the augmentation strategy, to deal with any molecular images that may appear in the real literature.
Please clone the following repositories:
git clone https://github.com/CYF2000127/MolNexTR
pip install -r requirements.txt
For training and inference, please download the following datasets to your own path.
- Synthetic: Indigo, ChemDraw
- Realistic: CLEF, UOB, USPTO, Staker, ACS
- Perturbed by img transform: CLEF, UOB, USPTO, Staker, ACS
- Perturbed by curved arrows: CLEF, UOB, USPTO, Staker, ACS
Run the following command:
sh ./exps/train.sh
The default batch size was set to 256. And it takes about 20 hours to train with 10 NVIDIA RTX 3090 GPUs.
Run the following command:
sh ./exps/eval.sh
The default batch size was set to 32 with a single NVIDIA RTX 3090 GPU.
Use prediction.ipynb for single or batched prediction and visualization.
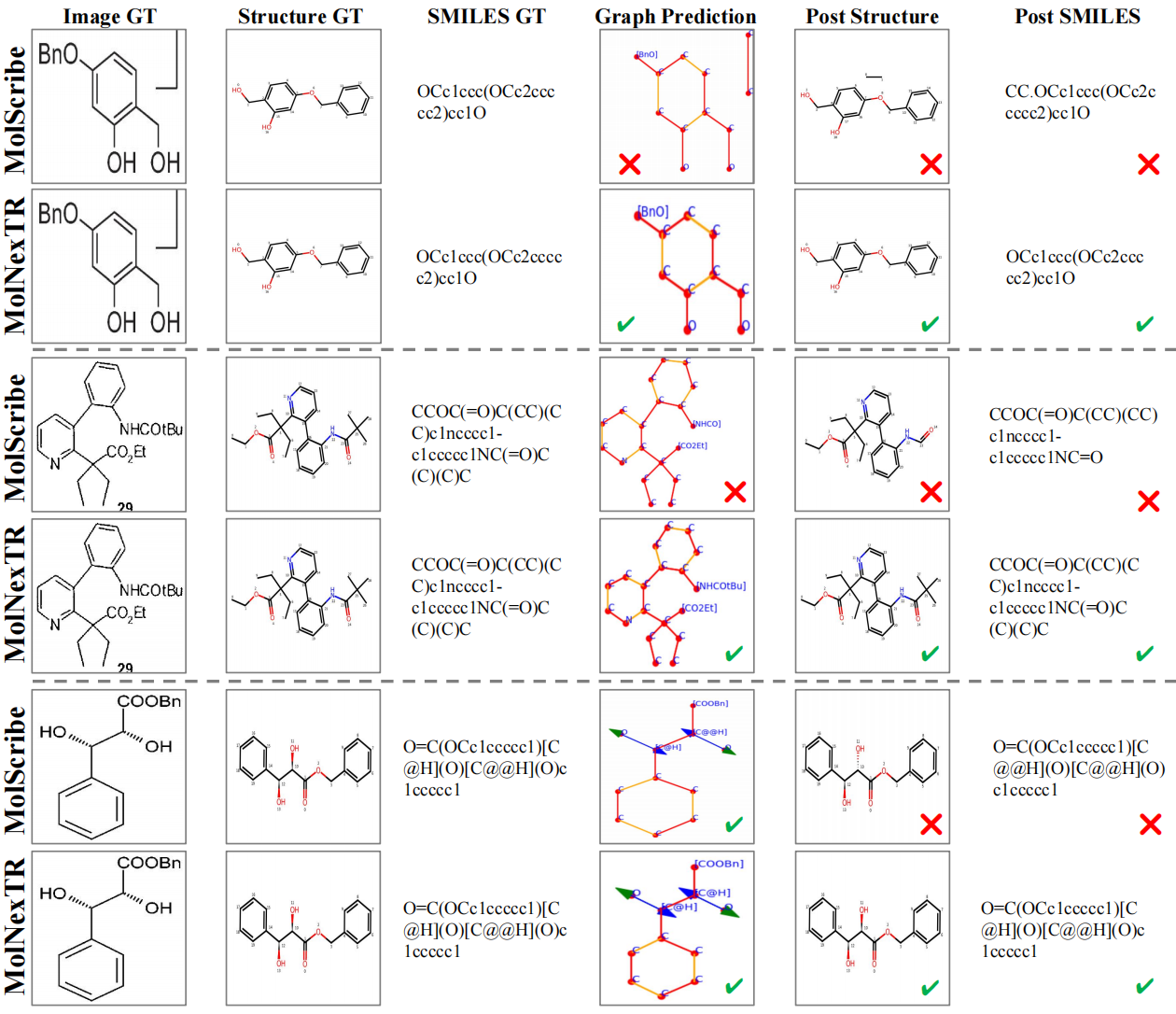
We also show some qualitative results of our method below: