- Authors: Chenhao Li, Tamar V. Av-Shalom, Jun Wei Gerald Tan, Junmei Samantha Kwah, Kern Rei Chng and Niranjan Nagarajan
BEEM-Static is an R package for learning directed microbial interactions from cross-sectional microbiome profiling data based on the generalized Lotka-Volterra model (gLVM). Extending the core idea of the original BEEM algorithm for longitudinal data (Reference, Source code), BEEM-Static directly works with relative abundances to jointly estimate total biomass and gLVM parameters, thus eliminating the need for experimentally quantifying absolute abundances. BEEM-Static identifies microbiomes that are not at equilibrium states and automatically excludes such samples from the analysis. The package also provides the user with a collection of utility functions for visualizing and diagnosing the fitted model.
Note: This package is under active development. Please record the commit ID for reproducibility.
devtools::install_github('lch14forever/BEEM-static')
library(beemStatic)The demo dataset is a simulated community of 20 species and 500 samples. All of the samples are at quilibrium states (generated by numerically integrating the gLVM until convergence) and each sample contains 70% of species as being randomly selected (i.e. each species has 70% habitat preference).
data("beemDemo")
attach(beemDemo)
## Use `?beemDemo` to see the help for fields in this datasetBEEM-Static is run by calling the func.EM function.
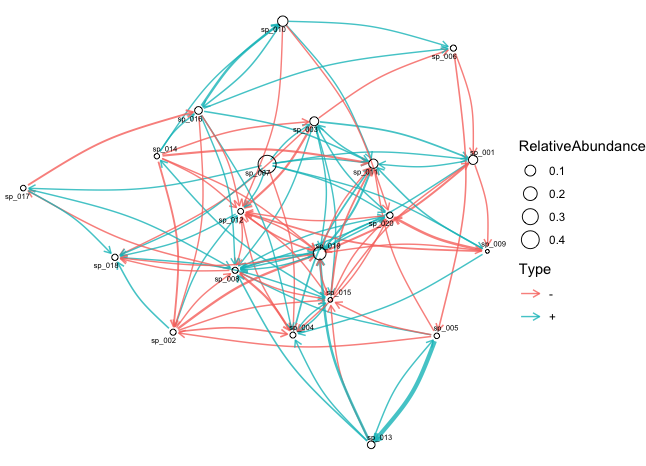
res <- func.EM(dat.w.noise, ncpu=4, scaling=median(biomass.true))We provide a function showInteraction to plot the interaction network inferred by BEEM-Static (based on the ggraph package).
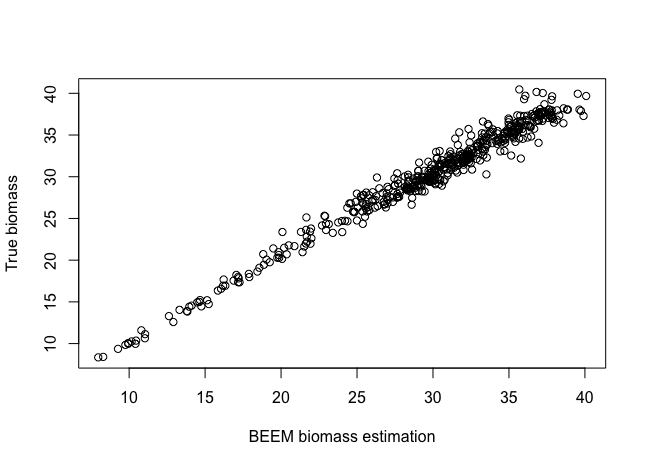
showInteraction(res, dat.w.noise)BEEM-Static also estimates the biomass for each sample (retrieved by the beem2biomass function). Here we can compare the estimated biomass with the true biomass on this simulated dataset.
plot(beem2biomass(res), biomass.true, xlab='BEEM biomass estimation', ylab='True biomass')We provide a function diagnoseFit to plot the coefficient of determination (R2) for each species. A high R2 (close to 1) value indicates that the variation in the data is well explained by the model.
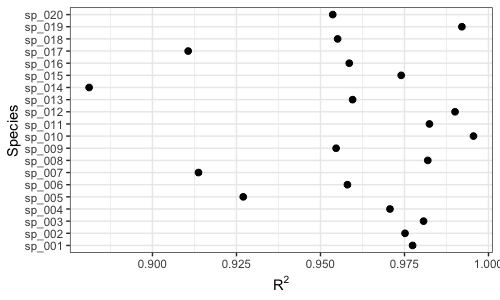
diagnoseFit(res, dat.w.noise, annotate = FALSE)We now run two popular methods for inferring microbial interactions on our simulated data. Both methods try to infer a correlation matrix as a proxy for the interaction matrix.
- Using a naive Spearman correlation method
spearman <- cor(t(dat.w.noise), method='spearman')- Using SPIEC-EASI
## devtools::install_github("zdk123/SpiecEasi")
library(SpiecEasi)
se <- spiec.easi(t(dat.w.noise), method='mb')
se.stab <- as.matrix(getOptMerge(se))- Using BEEM-Static
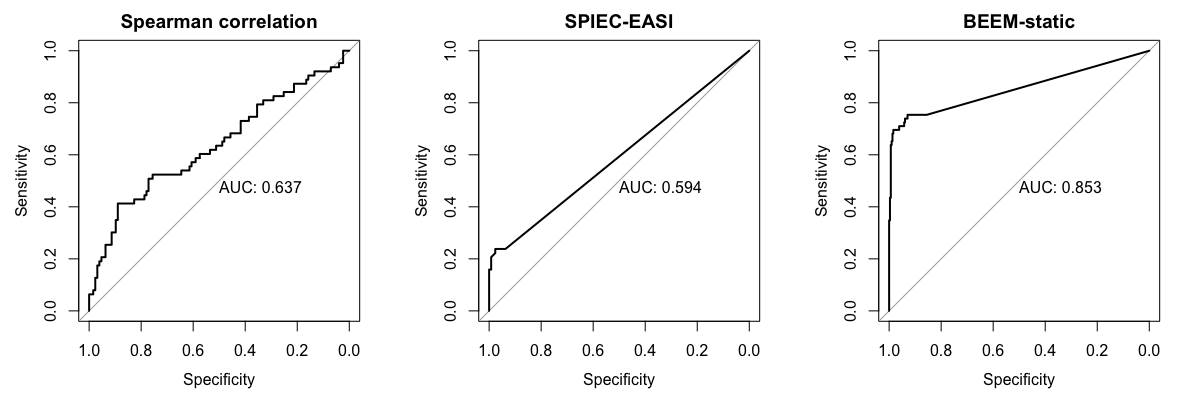
est <- beem2param(res)We implement a function auc.b for ploting the receiver operating characteristic (ROC) curve with computed area under the curve (AUC). We compare the ROC curves for the above three methods.
par(mfrow=c(1,3))
auc.b(spearman, scaled.params$b.truth, is.association = TRUE, main='Spearman correlation')
auc.b(se.stab, scaled.params$b.truth, is.association = TRUE, main='SPIEC-EASI')
auc.b(est$b.est, scaled.params$b.truth, main='BEEM-static')-
C Li, T V Av-Shalom et al. BEEM-Static: Accurate inference of ecological interactions from cross-sectional metagenomic data. bioRxiv 2020.
-
C Li, K R Chng, J S Kwah, T V Av-Shalom, L Tucker-Kellogg, N Nagarajan. An expectation-maximization algorithm enables accurate ecological modeling using longitudinal metagenome sequencing data. Microbiome 2019.
Please direct any questions or feeback to Li Chenhao and Niranjan Nagarajan.