PROT-ON’s primary aim is to deliver the critical (designer) PPI mutations that can be used to propose new protein binders. For this, PROT-ON uses the coordinates of a protein complex. It then probes all possible interface mutations with either EvoEF1 or FoldX on the selected protein monomer. The probed mutational landscape is then filtered according to the stability and optionally to the mutability criteria. PROT-ON finally statistically analyzes the energy landscape spanned by the probed mutation set with the aim of proposing the most binding enriching and depleting interfacial mutations.
This site describes the use of stand-alone version of PROT-ON, which is tested on Linux and MacOS systems. If you would like to use our tool through our web service, please visit http://proton.tools.ibg.edu.tr:8001
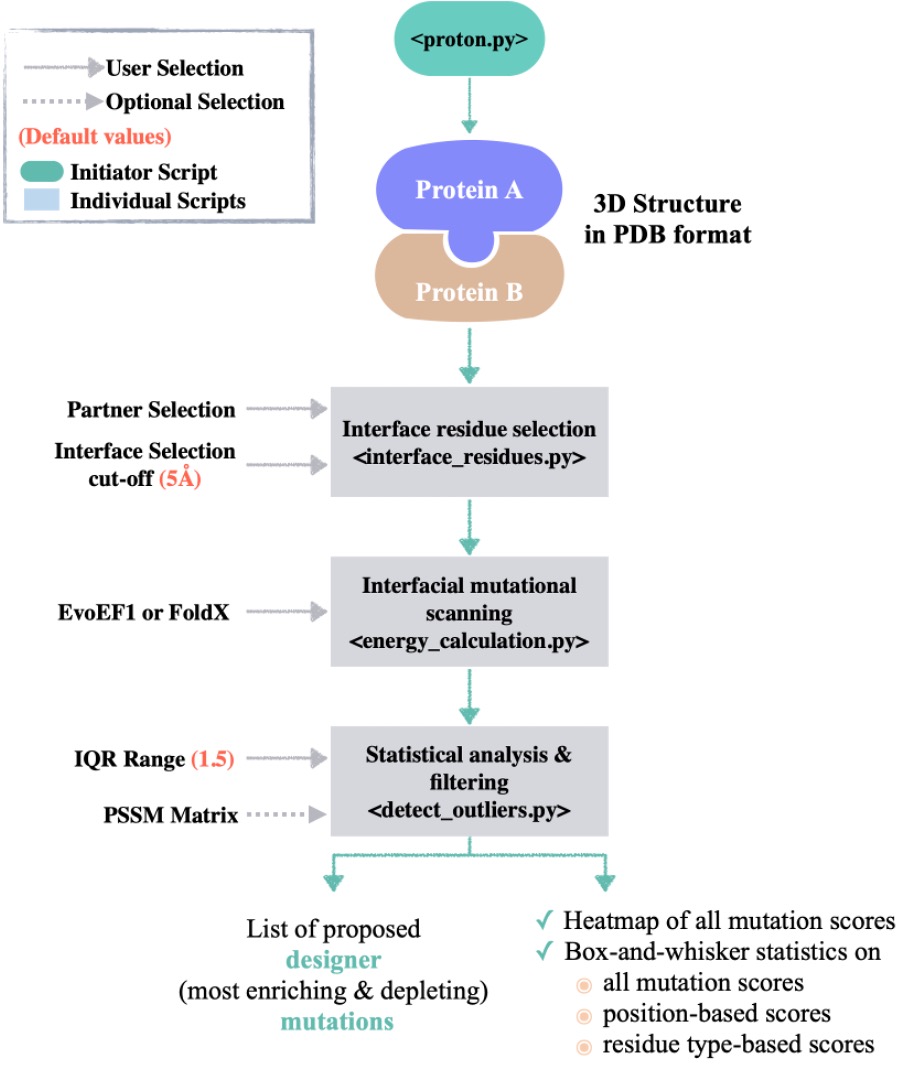
PROT-ON calls several python scripts to find the designer mutations (binding enriching/depleting) on one monomer of a complex. Here is the flow of the approach (pssm option is available only on the web server version):
- python3 OR conda (version 4.10 or higher)
- FoldX 4.0 (optional)
- numpy
- pandas (should be version 1.3.0 or higher)
- plotly
- kaleido
- shutil
- time
- virtualenv
git clone https://github.com/CSB-KaracaLab/prot-on.git
cd prot-on
We strongly suggest creating a Python Virtual Envrionment and activating it before the installation of PROT-ON dependencies. For this, please follow:
python3 -m venv <environment-name-you-choose>
source <environment-name-you-choose>/bin/activate
When the environment is active, please execute:
python setup.py
As a result of running setup.py, PROT-ON will be ready to perform the mutational scan with EvoEF1 (January-2021 version of EvoEF1 comes in the PROT-ON package).
-
Optional: If you would like to perform the mutational scanning with FoldX, you first have to get its academic licenced soure files. Among the obtained source files, please locate the FoldX executable (please name it as
foldx) androtabase.txtin theprot-on/srcfolder. Also, please make sure thatfoldxis executable in your system. -
Optional: If you would like to include the evolutionary information into the filtering process, please obtain the PSSM file of the monomer you will be scanning in the csv format. The external PSSM file, which can be obtained via https://possum.erc.monash.edu/server.jsp should be in a comma
,seperated format. Please name your PSSM file as<root-pdb-filename>_chain_<chain_ID>_pssm.csvand place it in theprot-onfolder. You can find an example PSSM file underexample-inputdirectory. -
After these steps, you can execute PROT-ON on your PDB formatted complex via:
python proton.py --pdb <dir/filename of the structure> --chain_ID <chain ID of interest> --algorithm <EvoEF1/FoldX> --cut_off <cut-off to define the interface> --IQR <IQR rule to define outliers of box-and-whisker statistics>
Example:
python proton.py --pdb example-input/complex.pdb --chain_ID D --algorithm EvoEF1 --cut_off 5.0 --IQR 1.5
cut-off, IQR and algorithm definitions are optional. By default they will be set to 5.0, 1.5, and EvoEF1 respectively. If these settings are fine with you, you can run PROT-ON with only:
python proton.py --pdb complex.pdb --chain_ID D
The help page of PROT-ON can be called with:
python proton.py --help
When PROT-ON is finished, your results will be located at results/runID_PDBID_chainID_output folder. In this folder, as given in the example-output folder, you will find:
- Parameters: Containing the defined interface cut-off and IQR range.
- Interface amino acid list: Interfacial amino acid list. This file corresponds to
complex_chain_D_interface_aa_listin theexample-outputfolder. - Pairwise inter-monomeric distance list:
complex_pairwise_distance_listas given in theexample-outputfolder. - Mutation list: The list of all possible interfacial mutations (format: KD28A; K: Wild-type amino acid, D: Chain ID, 28: Amino acid position, A: Mutant amino acid).
complex_chain_D_mutation_listas given in theexample-outputfolder. - Mutation models: The folder containing all the generated mutant structures.
- All EvoEF1/FoldX binding affinity predictions are analyzed with the box-whisker statistics, where;
- Depleting mutations: are defined by the positive outliers (as in
complex_chain_D_depleting_mutationsinexample-output), and; - Enriching mutations: are defined by the negative outliers (as in
complex_chain_D_enriching_mutationsinexample-output). - Heatmap source file: is given in
heatmap_dfin a single column format. This file can be converted ınto a 2x2 matrix by executingpivot_table = heatmap_df.pivot("Positions","Mutations","DDG_{}_Scores".format(algorithm) - Designer mutations: Stability-filtered enriching and depleting mutations, named as
complex_chain_D_stabilizing_enriching_mutationsandcomplex_chain_D_stabilizing_depleting_mutationsas given in theexample-outputdirectory . - Complete list of all PROT-ON scores: All the scores calculated throughout a single run are saved under
complex_chain_{}_proton_scores. - Graphical Outputs: We provide png and svg files of the scanned mutational energies as a heatmap, as a boxplot distribution representing all interfacial binding scores, as well as as boxplot distributions of the scores according to residue position and type.
Thank you for following our guidelines until this point! We hope that you will enjoy using our package!
We would like to thank Ayşe Berçin Barlas for her assistance in revising the code architecture.
If you use the PROT-ON, please cite the following paper:
Koşaca, M., Yılmazbilek, İ., & Karaca, E. (2023).
PROT-ON: A structure-based detection of designer PROTein interface MutatiONs.
Frontiers in Molecular Biosciences, 10, 1063971.
If you encounter any problem, you can contact Mehdi or Ezgi via: