Created by: Yazdan Asgari
Creation date: 14 Jan 2022
Update: Feb 2024
https://cesp.inserm.fr/en/equipe/exposome-and-heredity
NOTE: All SNPs and genes positions in the GWAS and annotation data used in the examples are based on GRCh37 (hg19) Human Genome Assembly.
-
Running the Pipeline using Shiny App (recommended for non-computer scientists) which is a R Shiny App that a user could easily open a Shiny App and change any desired parameters and run the whole procedure automatically with some simple clicks.
IMPORTANT NOTE: The Shiny App was tested on a Windows OS and Unix-based server with CentOS 7. -
Running the Pipeline using Bash File or R script (recommended for computer scientists) which is a plain text file that contains a series of commands for running the whole procedure automatically with a user-defined parameters.
IMPORTANT NOTE: The Bash file was tested on a Unix-based server with CentOS 7 while the R Script was tested on a Windows OS and Unix-based server with CentOS 7. -
Tutorial - Wiki which includes description for each scripts in more detail. This is useful for developers who want to modify/add any part of the pipeline
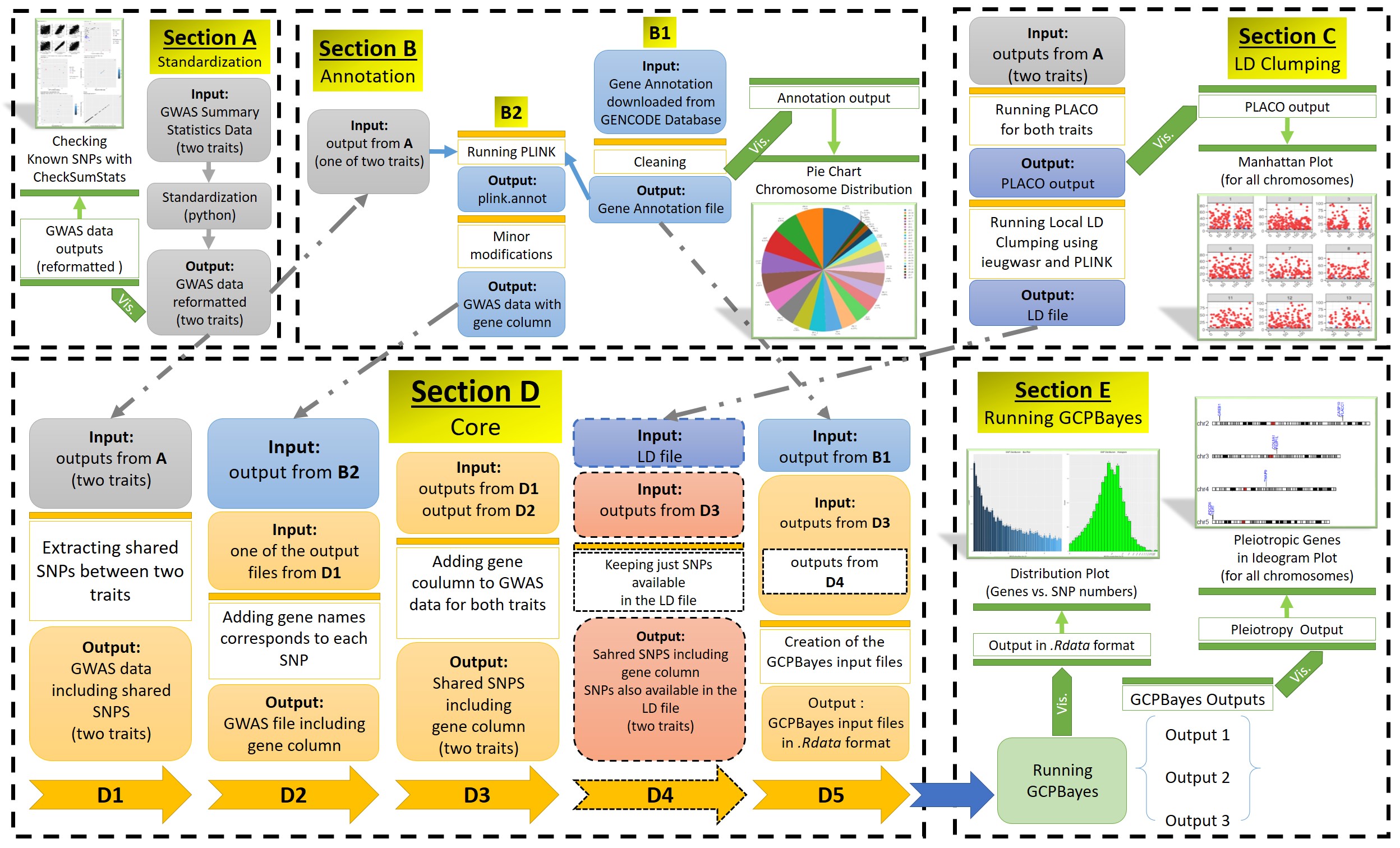
A schematic overview of the main sections of the GCPBayes pipeline is as follows:
Here, we provide a small dataset for testing the Pipeline. The data are GWAS summary statistics for The Breast Cancer Association Consortium (BCAC) and The Ovarian Cancer Association Consortium (OCAC) chromosome #5 and we want to run the Pipeline (without LD clumping method) and GCPBayes at a gene-level for 300 coding genes.
You could use one of the following options (R or Bash) to run a small example file to test GCPBayes Pipeline:
- Download and run the "GCPBayes_pipeline_check_packages_test.R" (Link) R script to check a list of required packages and install them if they are not available in your system. It also prints a warning message if any of the packages cannot be installed.
- Download INPUT files (Download)
- BCAC and OCAC GWAS data on chromosome #5 (gwas_BCAC_chr5.txt, gwas_OCAC_chr5.txt)
- An annotation file including all coding genes (annot_gencode_v38lift37_modified_gene_class.txt)
- BCAC GWAS file with a gene column (Annot_BCAC_2020_onco_ALL_reformatted_coding.txt)
- Download the scripts and put them in the same folder as input data (Download)
- R_C1_code_find_common_snps_one_pair_test.R
- R_D1_code_pipeline_annot_coding_withoutldclumping_extra_info_test.R
- R_D3_code_separate_groups_length_threshold_noclump_test.R
- R_E1_code_gcpbayes_less_extra_info_test.R
- Download the parameter file (GCPBayes_pipeline_parameters_test.R) (Download)
- You JUST need to replace /PATH/ regarding "working directory" with the path where you put all downloaded data and scripts.
- Now, all you need is to run "GCPBayes_pipeline_test.R" R script (Download)
- Download and run the "GCPBayes_pipeline_check_packages_test.R" (Link) R script to check a list of required packages and install them if they are not available in your system. It also prints a warning message if any of the packages cannot be installed.
- Download INPUT files (Download)
- BCAC and OCAC GWAS data on chromosome #5 (gwas_BCAC_chr5.txt, gwas_OCAC_chr5.txt)
- An annotation file including all coding genes (annot_gencode_v38lift37_modified_gene_class.txt)
- BCAC GWAS file with a gene column (Annot_BCAC_2020_onco_ALL_reformatted_coding.txt)
- Download the scripts and put them in the same folder as input data (Download)
- C1_code_find_common_snps_one_pair.R
- D1_code_pipeline_annot_coding_withoutldclumping_extra_info.R
- E1_code_gcpbayes_less_extra_info.R
- Download the parameter file (parameters_Strategy_bcac_ocac_test_set.ini) (Download)
- You MUST change this file before running. So, replace /PATH/ with the path where you put all downloaded data and scripts. You need to change these three parts:
- working directory
- output directory
- directory for scripts
- You MUST change this file before running. So, replace /PATH/ with the path where you put all downloaded data and scripts. You need to change these three parts:
- Download the readinput file (readinputs.txt) (Download)
- You MUST change replace /PATH/ with the same one you entered for the parameter file.
- Download the BASH file (run_test_set.sh) (Download)
- Now, all you need is to run the following command in the terminal:
$ ./run_test_set.sh parameters_Strategy_bcac_ocac_test_set.ini readinputs.txt
NOTE: You might need to change the permission of the BASH file in order to be executed.
$ chmod 777 run_test_set.sh
- step1_output_BCAC.txt
- step1_output_OCAC.txt
- D1_output_pipeline_BCAC_common2all_2_studies_coding_wo_clumping.txt
- D1_output_pipeline_OCAC_common2all_2_studies_coding_wo_clumping.txt
- D1_output_pipeline_SNP_in_genes_output_pipeline_output_wo_clump.txt
- D1_Summary_SNP_in_genes_output_pipeline_BCAC_common2all_2_studies_coding_wo_clumping.txt
- D1_Summary_SNP_in_genes_output_pipeline_OCAC_common2all_2_studies_coding_wo_clumping.txt
- D1_Matrices_extra_info_output_pipeline_output_wo_clump.Rdata
- D1_Matrices_output_pipeline_output_wo_clump.Rdata
- output_output_GCPBayes_wo_clump_less_threshold_700_results.txt
- output_output_GCPBayes_wo_clump_less_threshold_700_pleiotropy.txt
Running Time: It took about 3 minutes to run the pipeline before GCPBayes (in a system with Intel Core i7 11th Gen 2.8 GHz with 16 GB RAM). For the GCPBayes, we just used the first 300 genes from the data and it took 3 minutes to run.
Note: While running GCPBayes, a user could check these two files to see the results:
- output_output_GCPBayes_wo_clump_less_threshold_700_results.txt
- output_output_GCPBayes_wo_clump_less_threshold_700_pleiotropy.txt
Note: After a successful running, there would be a gene "SETD9" in the pleiotropic output file which determines the gene as a candidate with potential pleitropic signal among both breast and ovarian cancers. SETD9 has 43 SNPs through our test datasets and one of its biological functions is Regulation of TP53 Activity through Methylation.
- There are different visualizations for outputs from various steps. You can find more details through the visualization scripts in the Tutorial - Wiki.
- Shiny App - Online: For a GCPBayes pleiotropic candidate genes output, we developed a shiny App which you can find through the following link:
GCPBayes_Output_Shiny_App
For example, you could use this file as an example (Download) (filename: output_GCPBayes_pleiotropy_example.txt) (it has the same format as an output of GCPBayes pipeline for pleiotropic genes) and see different visualization tools via the online shiny App. NOTE: You need to select Space as separator after uploading the data.
- Shiny App - Local: It is also possible to use the script for the shiny App and run it on your computer. You could download the script from here (filename: shiny_gcpbayes_output.R). NOTE: You need to install the following packages before running the shiny App: shiny, datasets, ggplot2, gridExtra, tidyverse, BioCircos, plotly, and ggpubr.
- Shiny App with Karyotype - Local: For a newer version of the shiny App, we added a new graph (Karyotype) which demonstrates the position of candidate pleiotropic genes in the chromosomes. This type of graph is not available in the online version, but you could use it by running the shiny script on your computer. You could download the script from here (filename: shiny_gcpbayes_output_with_karyotype.R). NOTE: You need to install the following packages before running the shiny App: shiny, datasets, ggplot2, gridExtra, tidyverse, BioCircos, plotly, ggpubr, biomaRt, regioneR, and karyoploteR.
Asgari et al., "GCPBayes Pipeline: a tool for exploring pleiotropy at gene-level", NAR Genomics and Bioinformatics, 5(3), lqad065, 2023, doi:10.1093/nargab/lqad065 (Link)