- python>=3.7
- numpy
- scipy>=1.11.0
- cclib>=1.7.1 (If users want to use Gaussian …)
- matplotlib (If users want to obtain the figure of a profile)
Before running pyMCD, you should well configure the PYTHONPATH and the QC software to run pyMCD. For the latter one, you should configure the path as:
>> export PYTHONPATH=<git clone path>Also, QC software should be directly executable with a specific command. For example, in the case of Gaussian 16, when you command ‘which g16', the path for g16 should be found as the following:
>> which g16
>> /appl/Gaussian16/AVX_revB01/g16/g16If the path is not found, you will see the following result:
>> which g16
>> **no g16 in** (directories) # (No g16 is the key point)Users have to prepare three input files:
- R.com: Input for the geometry of reactants
- coordinates: Input for setting active coordinates
- qc_input: Input for Quantum Chemistry (QC) package
- options: Additional hyperparameters for pyMCD
Here, we show the example of input files in example/diels
The geometry of target molecule with charge and spin multiplicity is written:
0 1 # charge multiplicity
C 0.00000000 0.00000000 0.00000000 # 3D geometry of reactants of Diels alder reaction
C 0.03101500 0.00321900 1.33695700
C 1.25868100 0.00290300 2.13867800
C 2.49534700 -0.00205700 1.62972600
C 2.26606500 -2.42640100 0.45058000
C 1.15192300 -2.42622200 -0.27662600
H 0.17926100 -2.67118800 0.12875300
H 1.12996800 -2.20777400 -1.33507000
H 2.28637300 -2.67106300 1.50421500
H 3.24383300 -2.20776500 0.04480200
H 3.38321900 -0.04383300 2.24341300
H 2.71059800 0.07480700 0.57265500
H 1.10020000 -0.03373600 3.21984800
H -0.89519400 -0.03471900 1.91664600
H -0.91872700 -0.04169400 -0.56646200
H 0.88142600 0.07857400 -0.62177800
Active coordinates in terms of internal coordinates are specified
1 6 1.54 6
4 5 1.54 6
Here, it scans the distance between C1 and C6 and the distance between C4 and C5. They are shrunk to 1.54 angstrom with 6 steps.
Information containing theory level of calculations, basis set, solvation, and other detail options.
For gaussian,
#N pm6 scf(xqc)
For Orca,
! b3lyp 6-31g(d)
We feed hyperparameters of pyMCD as the following format:
num_relaxation=7 # The maximum number of optimization steps, default=5
step_size=0.05 # The maximal displacement within an optimization step, default is set with optimized value
unit=Hartree # The energy unit for printing a log file. Available units are kcal, eV, Hartree. Default is Hartree
working_directory=<scratch directory> # A scratch directory for running QC package, default is the input directory
calculator= Gaussian # The name of QC package. Currently, Gaussian and Orac are available
command=g09 # The command for running QC package. (Same command you configured in 2. Settings)
use_hessian=0 # Whether to use the original version or the revised version of MCD
hessian_update=exact # Method for calculating the Hessian
reoptimize=1 # Whether to first optimize the given reactant geometry before running the MCD algorithmThis file is not necessary. If this file is not found, the code will automatically run with setting variables as default values.
You can run pyMCD with the following command:
python run.py -id <input directory> -od <output directory>Output files will be saved in the output directory. Default option is same with the input directory
Example Code for diels-alder reaction TS search
python run.py \
-id example/diels \
-od example/diels
Basically, five output files are created:
- output.log
- pathway.xyz
- ts.xyz
- profile.log
- profile.png (This is well created only when the matplotlib library is well installed)
Example output files after running pyMCD with the previous input are shown below:
A brief description and output logs of pyMCD are written.
###### Reactant information ######
charge: 0 multiplicity: 1
16
C 0.0 0.0 0.0
C 0.031015 0.003219 1.336957
C 1.258681 0.002903 2.138678
C 2.495347 -0.002057 1.629726
C 2.266065 -2.426401 0.45058
C 1.151923 -2.426222 -0.276626
H 0.179261 -2.671188 0.128753
H 1.129968 -2.207774 -1.33507
H 2.286373 -2.671063 1.504215
H 3.243833 -2.207765 0.044802
H 3.383219 -0.043833 2.243413
H 2.710598 0.074807 0.572655
H 1.1002 -0.033736 3.219848
H -0.895194 -0.034719 1.916646
H -0.918727 -0.041694 -0.566462
H 0.881426 0.078574 -0.621778
######### Scanning coordinates #########
(1, 6): 2.7 -> 1.54, -0.1933 angstrom per step
(4, 5): 2.7056 -> 1.54, -0.1943 angstrom per step
Step size is set to 0.1166 as the default value!!!
########## Scanning information ##########
num relaxation: 5
step size: 0.1166
##### Calculator information ######
Calculator: Gaussian
working_directory: /scratch/udg/
command: g16
Energy: Hartree
###### qc_input ######
#N pm6 scf(xqc)
Starting time: 2022-10-21 11:45:45.372045
Set up done!!!
Initialization, Relaxing ....
Relaxation finished! Start scanning ....
[2022-10-21 11:45:48.417] Progress (1/12): (1, 6): 0/6 (4, 5): 1/6
[2022-10-21 11:46:02.254] 0.3921E-2 Hartree has Increased after 5 force calls! 0:00:15.312 Taken ...
[2022-10-21 11:46:03.746] Progress (2/12): (1, 6): 1/6 (4, 5): 1/6
[2022-10-21 11:46:14.718] 0.4016E-2 Hartree has Increased after 5 force calls! 0:00:12.460 Taken ...
[2022-10-21 11:46:16.204] Progress (3/12): (1, 6): 2/6 (4, 5): 1/6
...
[2022-10-21 11:48:10.626] Progress (12/12): (1, 6): 6/6 (4, 5): 6/6
[2022-10-21 11:48:16.122] 0.1393E-1 Hartree has Decreased after 5 force calls! 0:00:05.497 Taken ...
[2022-10-21 11:48:16.126018] Scan completed ...
Total 60 force calls performed ...
End time: 2022-10-21 11:48:17.883163
Taken time: 0:02:32.511118
The geometries of the points along the pathway generated by pyMCD ant their electronic energies are written.
0
0.0776422942853723
C 0.0 0.0 0.0
C 0.031015 0.003219 1.336957
C 1.258681 0.002903 2.138678
C 2.495347 -0.002057 1.629726
C 2.266065 -2.426401 0.45058
C 1.151923 -2.426222 -0.276626
H 0.179261 -2.671188 0.128753
H 1.129968 -2.207774 -1.33507
H 2.286373 -2.671063 1.504215
H 3.243833 -2.207765 0.044802
H 3.383219 -0.043833 2.243413
H 2.710598 0.074807 0.572655
H 1.1002 -0.033736 3.219848
H -0.895194 -0.034719 1.916646
H -0.918727 -0.041694 -0.566462
H 0.881426 0.078574 -0.621778
...
12
-0.0018241595178008856
C 0.095939 -0.570428 -0.030593
C 0.107274 -0.044635 1.374394
C 1.244687 -0.017894 2.077553
C 2.507095 -0.551025 1.466278
C 2.22822 -1.860973 0.706101
C 1.004303 -1.798535 -0.226117
H 0.401402 -2.715241 -0.075825
H 1.340158 -1.827626 -1.279694
H 2.074158 -2.669841 1.447624
H 3.124858 -2.144978 0.125054
H 3.283953 -0.721542 2.233865
H 2.922535 0.206876 0.769674
H 1.311118 0.367402 3.088442
H -0.839231 0.316348 1.763824
H -0.934544 -0.820205 -0.34714
H 0.43206 0.245347 -0.707103
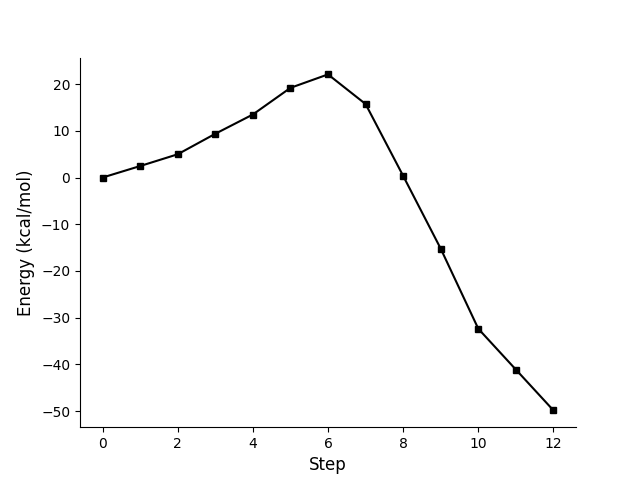
The electronic energies of the points along the pathway are written. The first column shows the original energy with the energy unit specified in the ‘option’ file. The second column shows the relative energy with respect to reactants with kcal/mol.
Original Energy (Hartree/mol) Relative Energy (kcal/mol)
0.0776422942853723 0.0
0.0815632928108389 2.460463722517743
0.08557910769066236 4.980425605690531
0.09257505477825809 9.370448683201001
0.09915399868185863 13.498798312040018
0.10824465842889286 19.20327342878806
0.11283916423722741 22.08636935215593
0.10272448736475942 15.739313787595341
0.07821446750017079 0.35904411309119594
0.05336164001392209 -15.236340591785684
0.02604045476074957 -32.380643180784496
0.012102670988113632 -41.12673454555648
-0.0018241595178008856 -49.86595263168884
Maxima point: 6
Figure of profile using relative energy as y-axis.
The geometry of the maximum point of the pathway and its electronic energy.
16
0.11283916423722741
C 0.118648 -0.215641 -0.044401
C 0.067769 -0.053241 1.321442
C 1.252311 -0.059302 2.09557
C 2.484097 -0.21923 1.503589
C 2.202348 -2.117006 0.595079
C 1.045665 -2.119159 -0.152426
H 0.14 -2.577299 0.225919
H 1.078937 -2.023226 -1.230977
H 2.233934 -2.570386 1.579193
H 3.172997 -2.011822 0.12569
H 3.373477 -0.385348 2.095592
H 2.696236 0.157142 0.506919
H 1.157129 -0.10527 3.180968
H -0.887984 -0.089442 1.843352
H -0.783879 -0.365275 -0.622262
H 0.952301 0.147552 -0.636911
View the following page: Implementation instruction
Please cite as below Lee, Kyunghoon, Jun Hyeong Kim, and Woo Youn Kim. "pyMCD: Python package for searching transition states via the multicoordinate driven method." Computer Physics Communications 291 (2023): 108831.
The code is under BSD-3-license
Please e-mail me to here: kyunghoonlee@kaist.ac.kr