This github repository contains the code used to achieve the results reported in MemoryInception: Predicting Neurological Recovery from EEG Using Recurrent Inceptions. Full reference to the original paper :
B. -J. Singstad, J. Ravn and A. Ranjbar, "MemoryInception: Predicting Neurological Recovery from EEG Using Recurrent Inceptions," 2023 Computing in Cardiology (CinC), Atlanta, GA, USA, 2023, pp. 1-4, doi: 10.22489/CinC.2023.133.
Main branch - organizers baseline code
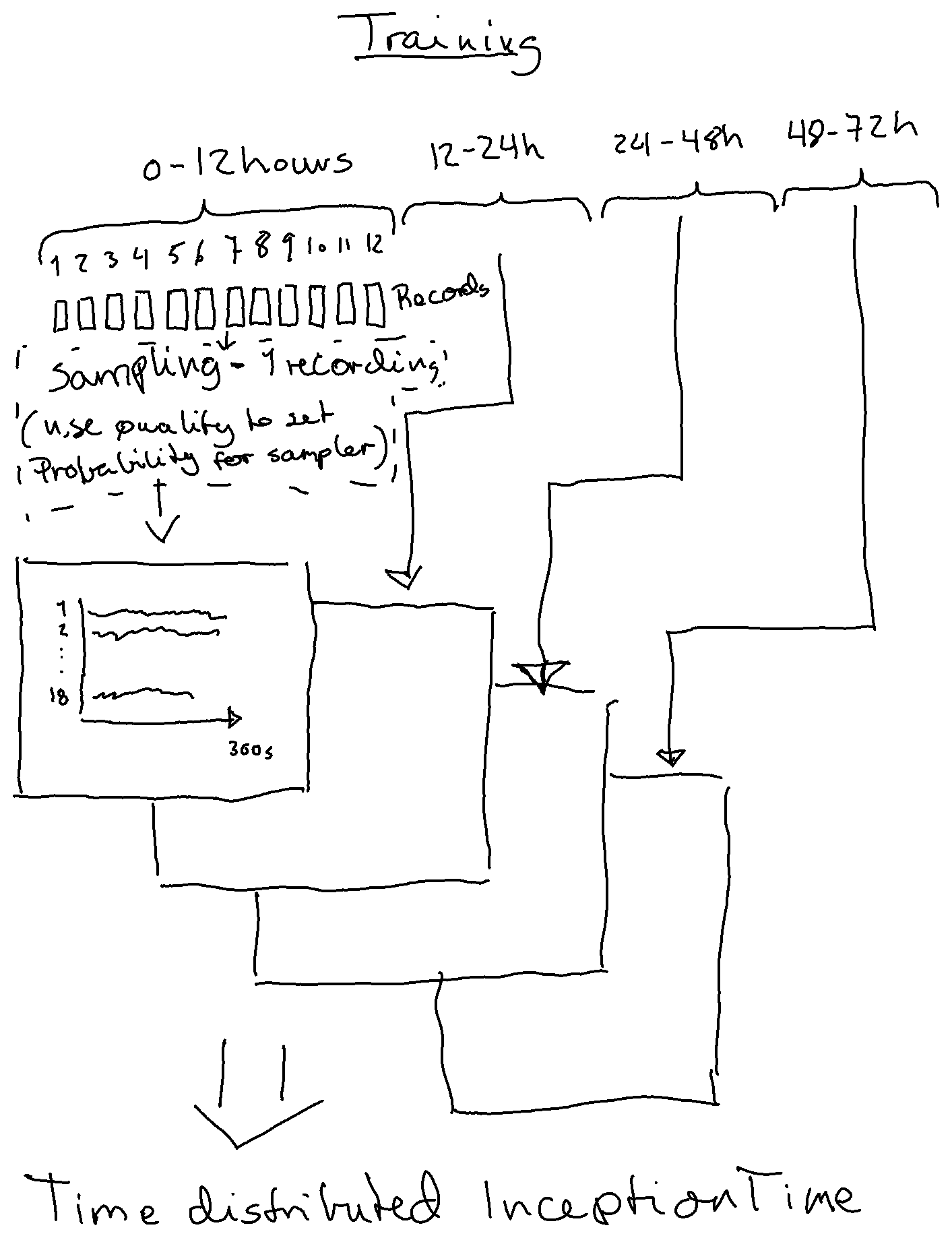
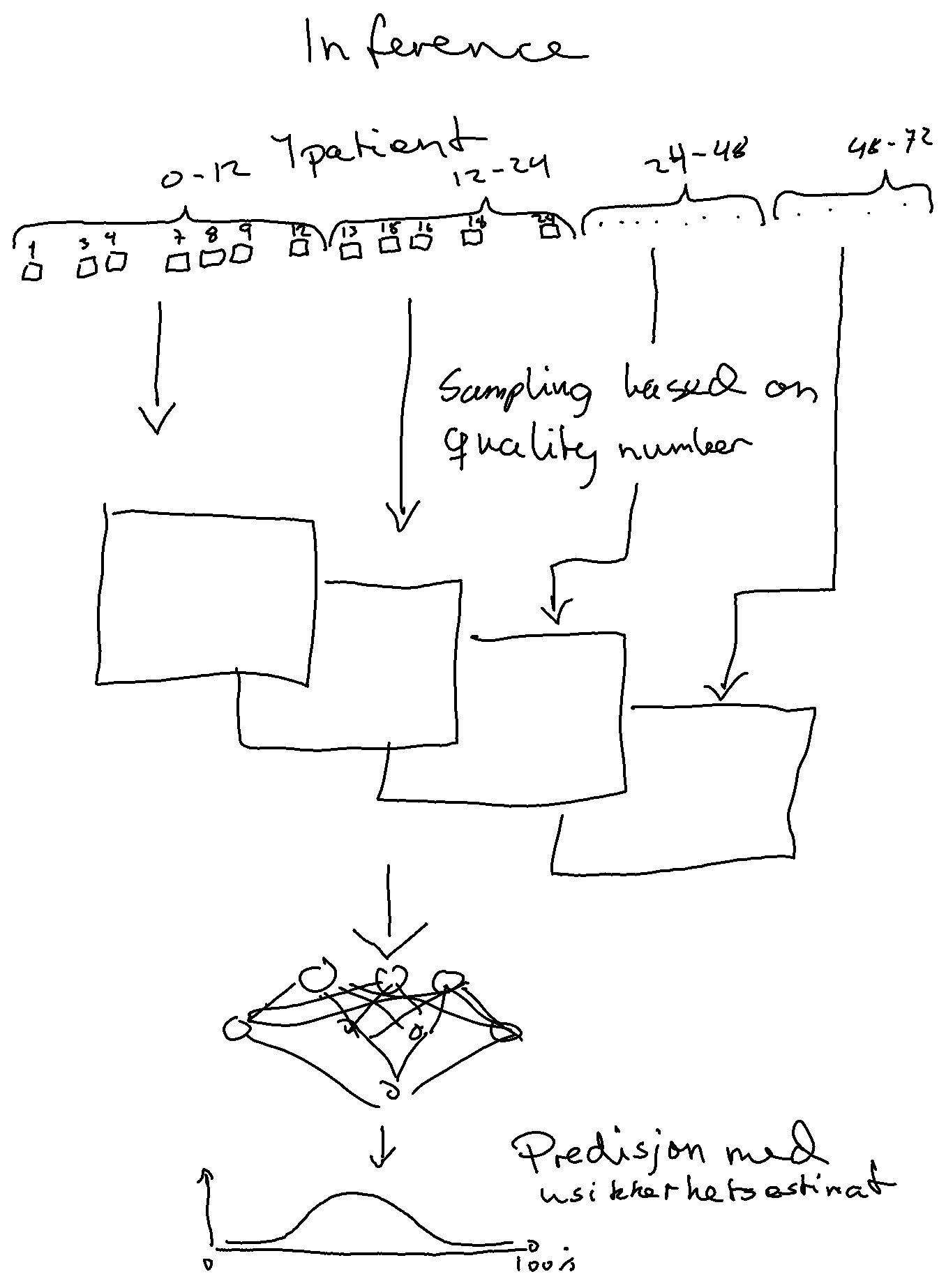
inception_time_dev branch - inception time using one 5min recording pr patient
This repository contains a simple example to illustrate how to format a Python entry for the George B. Moody PhysioNet Challenge 2023. You can try it by running the following commands on the Challenge training sets. These commands should take a few minutes or less to run from start to finish on a recent personal computer.
For this example, we implemented a random forest model with several features. You can use different models, features, and libraries for your entry. This simpple example is designed not not to perform well, so you should not use it as a baseline for your model's performance.
This code uses four main scripts, described below, to train and run a model for the Challenge.
You can install the dependencies for these scripts by creating a Docker image (see below) and running
pip install -r requirements.txt
You can train you model by running
python train_model.py training_data model
where
training_data(input; required) is a folder with the training data files andmodel(output; required) is a folder for saving your model.
You can run you trained model by running
python run_model.py model test_data test_outputs
where
model(input; required) is a folder for loading your model, andtest_data(input; required) is a folder with the validation or test data files (you can use the training data for debugging and cross-validation), andtest_outputsis a folder for saving your model outputs.
The Challenge website provides a training database with a description of the contents and structure of the data files.
You can evaluate your model by pulling or downloading the evaluation code and running
python evaluate_model.py labels outputs scores.csv
where labels is a folder with labels for the data, such as the training database on the PhysioNet webpage; outputs is a folder containing files with your model's outputs for the data; and scores.csv (optional) is a collection of scores for your model.
We will run the train_model.py and run_model.py scripts to train and run your model, so please check these scripts and the functions that they call.
Please edit the following script to add your training and testing code:
team_code.pyis a script with functions for training and running your model.
Please do not edit the following scripts. We will use the unedited versions of these scripts when running your code:
train_model.pyis a script for training your model.run_model.pyis a script for running your trained model.helper_code.pyis a script with helper functions that we used for our code. You are welcome to use them in your code.
These scripts must remain in the root path of your repository, but you can put other scripts and other files elsewhere in your repository.
To train and save your models, please edit the train_challenge_model function in the team_code.py script. Please do not edit the input or output arguments of the train_challenge_model function.
To load and run your trained model, please edit the load_challenge_model and run_challenge_model functions in the team_code.py script. Please do not edit the input or output arguments of the functions of the load_challenge_model and run_challenge_model functions.
Docker and similar platforms allow you to containerize and package your code with specific dependencies so that you can run your code reliably in other computing environments and operating systems.
To guarantee that we can run your code, please install Docker, build a Docker image from your code, and run it on the training data. To quickly check your code for bugs, you may want to run it on a small subset of the training data.
If you have trouble running your code, then please try the follow steps to run the example code.
-
Create a folder
examplein your home directory with several subfolders.user@computer:~$ cd ~/ user@computer:~$ mkdir example user@computer:~$ cd example user@computer:~/example$ mkdir training_data test_data model test_outputs -
Download the training data from the Challenge website. Put some of the training data in
training_dataandtest_data. You can use some of the training data to check your code (and should perform cross-validation on the training data to evaluate your algorithm). -
Download or clone this repository in your terminal.
user@computer:~/example$ git clone https://github.com/physionetchallenges/python-example-2023.git -
Build a Docker image and run the example code in your terminal.
user@computer:~/example$ ls model python-example-2023 test_data test_outputs training_data user@computer:~/example$ cd python-example-2023/ user@computer:~/example/python-example-2023$ docker build -t image . Sending build context to Docker daemon [...]kB [...] Successfully tagged image:latest user@computer:~/example/python-example-2023$ docker run -it -v ~/example/model:/challenge/model -v ~/example/test_data:/challenge/test_data -v ~/example/test_outputs:/challenge/test_outputs -v ~/example/training_data:/challenge/training_data image bash root@[...]:/challenge# ls Dockerfile README.md test_outputs evaluate_model.py requirements.txt training_data helper_code.py team_code.py train_model.py LICENSE run_model.py root@[...]:/challenge# python train_model.py training_data model root@[...]:/challenge# python run_model.py model test_data test_outputs root@[...]:/challenge# python evaluate_model.py test_data test_outputs [...] root@[...]:/challenge# exit Exit
Please see the Challenge website for more details. Please post questions and concerns on the Challenge discussion forum.