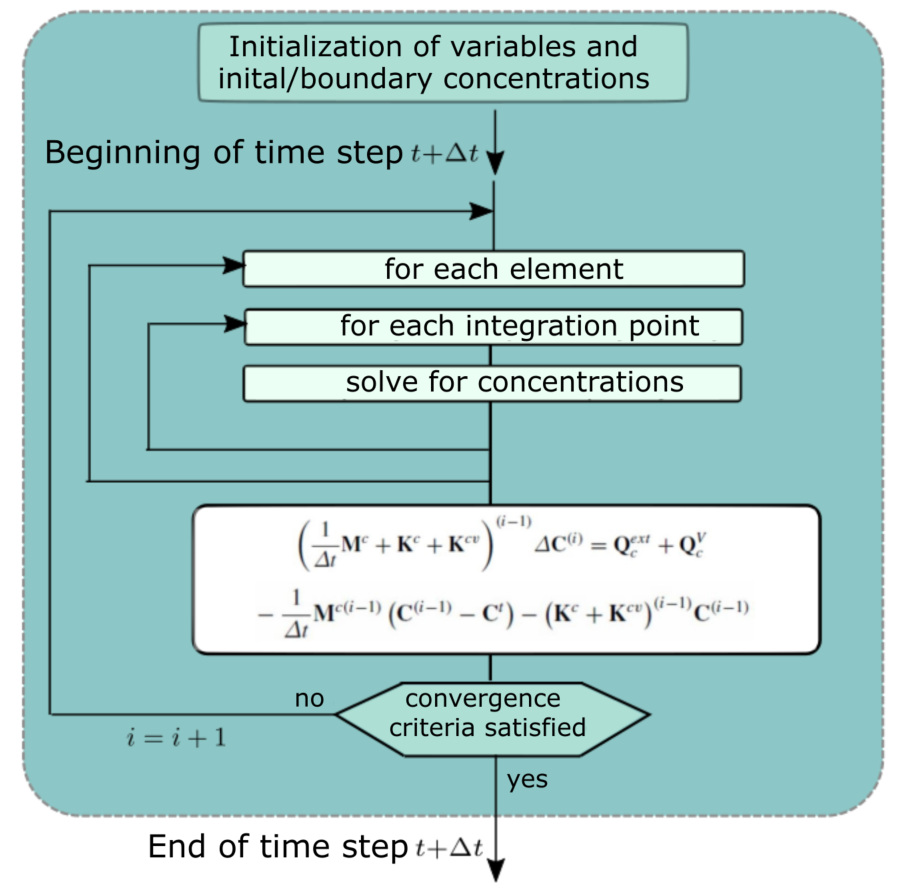
PAK-KTM (Program for Structural Analysis - Kojic Transport Model) is a high-performance software for finite element analysis (FEA) of gradient-driven smeared physical fields, developed at the University of Kragujevac and the Research and Development Center for Bioengineering, BioIRC. The Kojic Transport Model is based on the composite smeared finite element (CSFE) and is used for modelling complex multiscale transport problems in the biological composite media. The program is written in FORTRAN 77/90/95 and is capable of solving diffusion and convection fields, enabling the modelling of the mass transport of ions or molecules. Within CSFE, the detailed biochemical properties are included for the tissues as composite continuum media, as well as 1D structures and biological barriers at vessel walls and cell membranes. The fundamental laws utilized include Fick's law of diffusion, from which the mass balance equation is derived and translated into a form applicable to the finite element method. The model incorporates the assumption that the transport of molecules can be described by a nonlinear, non-stationary diffusion process.
The methodology regarding the KTM model is summarized in the book:
- M. Kojic, M. Milosevic, A. Ziemys, “Computational Models in Biomedical Engineering - Finite Element Models Based on Smeared Physical Fields: Theory, Solutions, and Software", Elsevier, 2022. [http://dx.doi.org/10.1016/j.cma.2015.06.002]
The software involves modeling systems for the analysis and assessment of convective-diffusive transport in complex systems such as tumors and organs consisting of a large number of capillaries and subdomains (capillary domain, lymphatic domain, tissue domain, cell domain, intercellular space domain, etc.). Determination of basic mass transport process variables such as velocities, pressure, or concentration is performed using the finite element method (FEM).
Modern medicine involves the application of the latest technologies for more efficient human treatment and predicting disease development, as well as predicting drug delivery within complex biological systems. As a result of numerical calculations using this method, significant information is obtained, along with a better understanding of transport processes occurring in the vascular system, organs, and intercellular space. Users of this technical solution can model complex tissue and organ systems in a very simple way, using distributed (smearedstrong>) fields, significantly facilitating the generation of complex systems and organs and further reducing costs for pre-clinical and clinical trials.
Another law employed is Darcy's law, which describes the transport of fluids in porous media through convection. When using this software, please cite the aforementioned book or the following papers:
-
M. Kojic, M. Milosevic, N. Kojic, Z. Starosolski, K. Ghaghada, R. Serda, A. Annapragada, M. Ferrari, and A. Ziemys, “A multi-scale FE model for convective–diffusive drug transport within tumor and large vascular networks,” Computer Methods in Applied Mechanics and Engineering, vol. 294. Elsevier BV, pp. 100–122, Sep-2015. [http://dx.doi.org/10.1016/j.cma.2015.06.002]
-
M. Kojic, M. Milosevic, N. Kojic, E. J. Koay, J. B. Fleming, M. Ferrari, and A. Ziemys, “Mass release curves as the constitutive curves for modeling diffusive transport within biological tissue,” Computers in Biology and Medicine, vol. 92. Elsevier BV, pp. 156–167, Jan-2018. [https://doi.org/10.1016/j.compbiomed.2016.06.026]
-
M. Kojic, M. Milosevic, V. Simic, E. J. Koay, J. B. Fleming, S. Nizzero, N. Kojic, A. Ziemys, and M. Ferrari, “A composite smeared finite element for mass transport in capillary systems and biological tissue,” Computer Methods in Applied Mechanics and Engineering, vol. 324. Elsevier BV, pp. 413–437, Sep-2017. [http://dx.doi.org/10.1016/j.cma.2017.06.019]
-
M. Kojić, M. Milošević, V. Simić, E. J. Koay, N. Kojić, A. Ziemys, and M. Ferrari, “EXTENSION OF THE COMPOSITE SMEARED FINITE ELEMENT (CSFE) TO INCLUDE LYMPHATIC SYSTEM IN MODELING MASS TRANSPORT IN CAPILLARY SYSTEMS AND BIOLOGICAL TISSUE,” Journal of the Serbian Society for Computational Mechanics, vol. 11, no. 2. Faculty of Engineering, University of Kragujevac, pp. 108–119, Dec-2017. [http://doi.org/10.24874/jsscm.2017.11.02.09]
-
M. Milosevic, V. Simic, B. Milicevic, E. J. Koay, M. Ferrari, A. Ziemys, and M. Kojic, “Correction function for accuracy improvement of the Composite Smeared Finite Element for diffusive transport in biological tissue systems,” Computer Methods in Applied Mechanics and Engineering, vol. 338. Elsevier BV, pp. 97–116, Aug-2018.[https://doi.org/10.1016/j.cma.2018.04.012]
-
M. Kojic, M. Milosevic, V. Simic, E. J. Koay, N. Kojic, A. Ziemys, and M. Ferrari, “Multiscale smeared finite element model for mass transport in biological tissue: From blood vessels to cells and cellular organelles,” Computers in Biology and Medicine, vol. 99. Elsevier BV, pp. 7–23, Aug-2018. [https://doi.org/10.1016/j.compbiomed.2018.05.022]
-
M. Milosevic, D. Stojanovic, V. Simic, B. Milicevic, A. Radisavljevic, P. Uskokovic, and M. Kojic, “A Computational Model for Drug Release from PLGA Implant,” Materials, vol. 11, no. 12. MDPI AG, p. 2416, 29-Nov-2018. [http://dx.doi.org/10.3390/ma11122416]
-
R. Santagiuliana, M. Milosevic, B. Milicevic, G. Sciumè, V. Simic, A. Ziemys, M. Kojic, and B. A. Schrefler, “Coupling tumor growth and bio distribution models,” Biomedical Microdevices, vol. 21, no. 2. Springer Science and Business Media LLC, 25-Mar-2019. [https://doi.org/10.1007/s10544-019-0368-y]
Data flow
Kojic transport element
Connective elements
-
Windows:
-
VisualStudio >=2017
-
Intel® oneAPI Base Toolkit
-
Intel® Distribution for GDB*
-
Intel® oneAPI HPC Toolkit
-
Linux:
-
GNU Fortran (GCC) 4.4.7 20120313
-
The C compiler GNU 4.4.7
-
MUMPS 5.0.2.
Manual and requirements can be viewed HERE. The file contains instructions on how to compile and run PAKKTM, details about the input and output files, along with result visualization instructions.
cmake ..
make
./pakktm
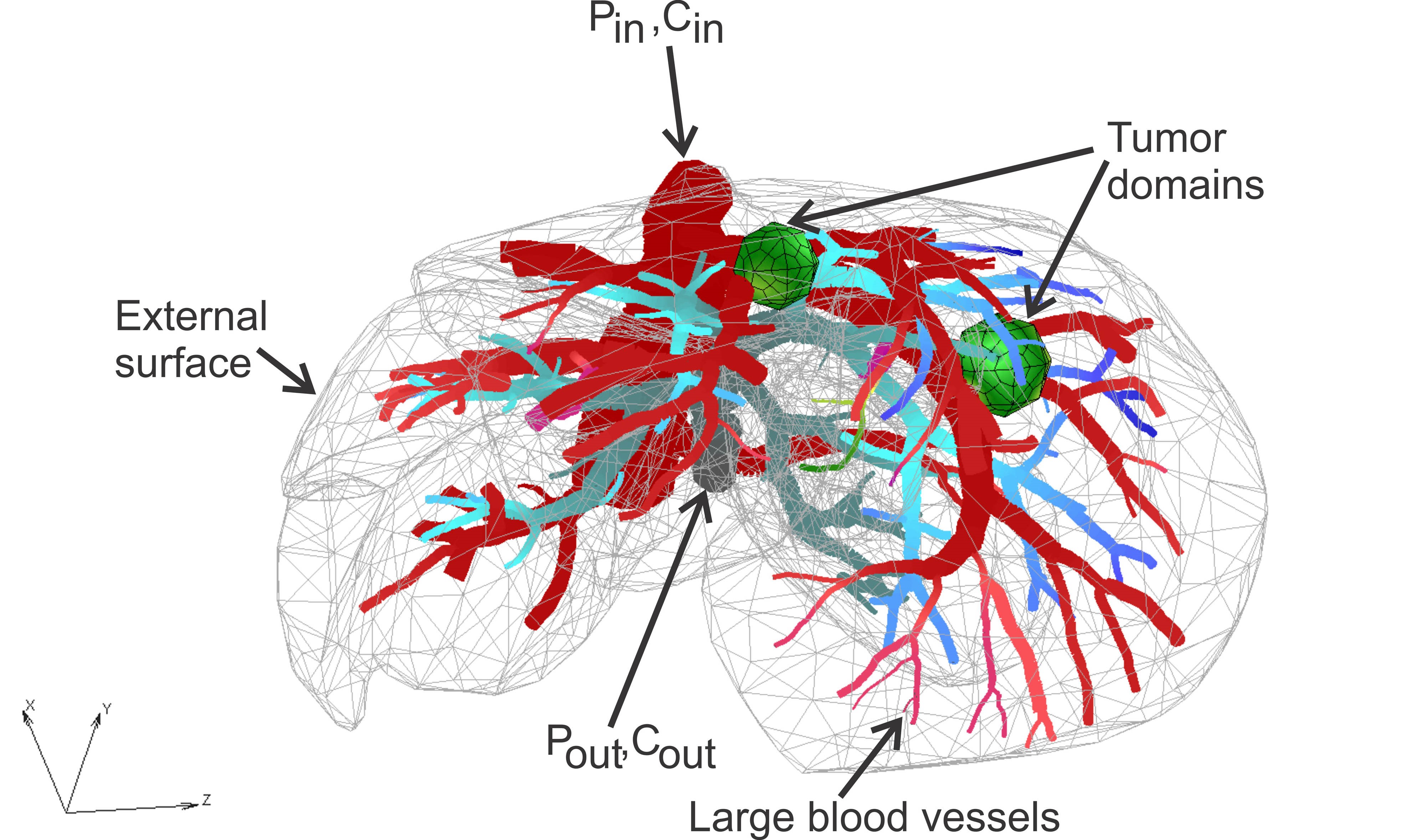
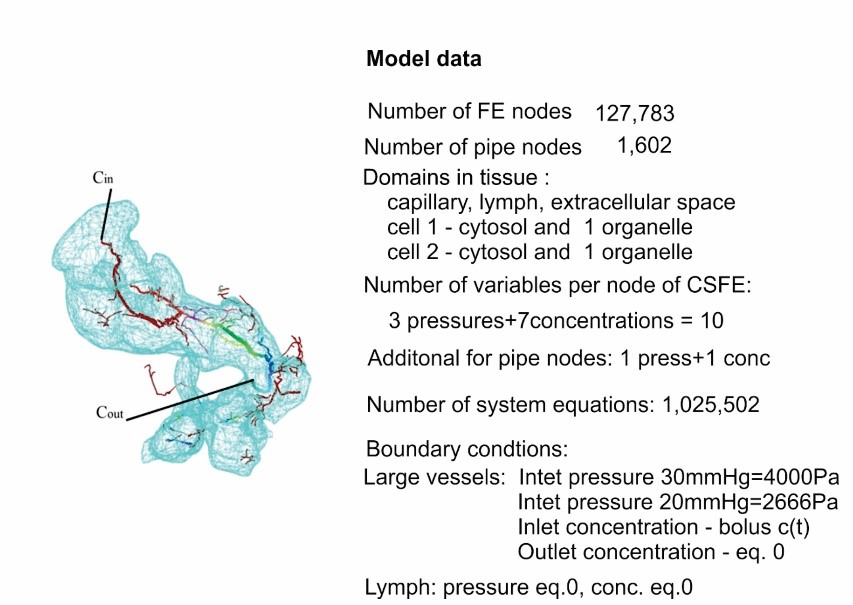
The finite element model consists of 1D pipe FEs for larger vessels (7736 elements), 3D composite smeared elements (39832 elements), and connectivity elements (726 elements) for connecting large vessels with continuum nodes (capillary domain DOF) of smeared FEs. There are two separate tumors within liver, with a total of 316 elements. The total number of nodes is 54590.
Liver model: geometry, tumor domains and pressures within large vessels
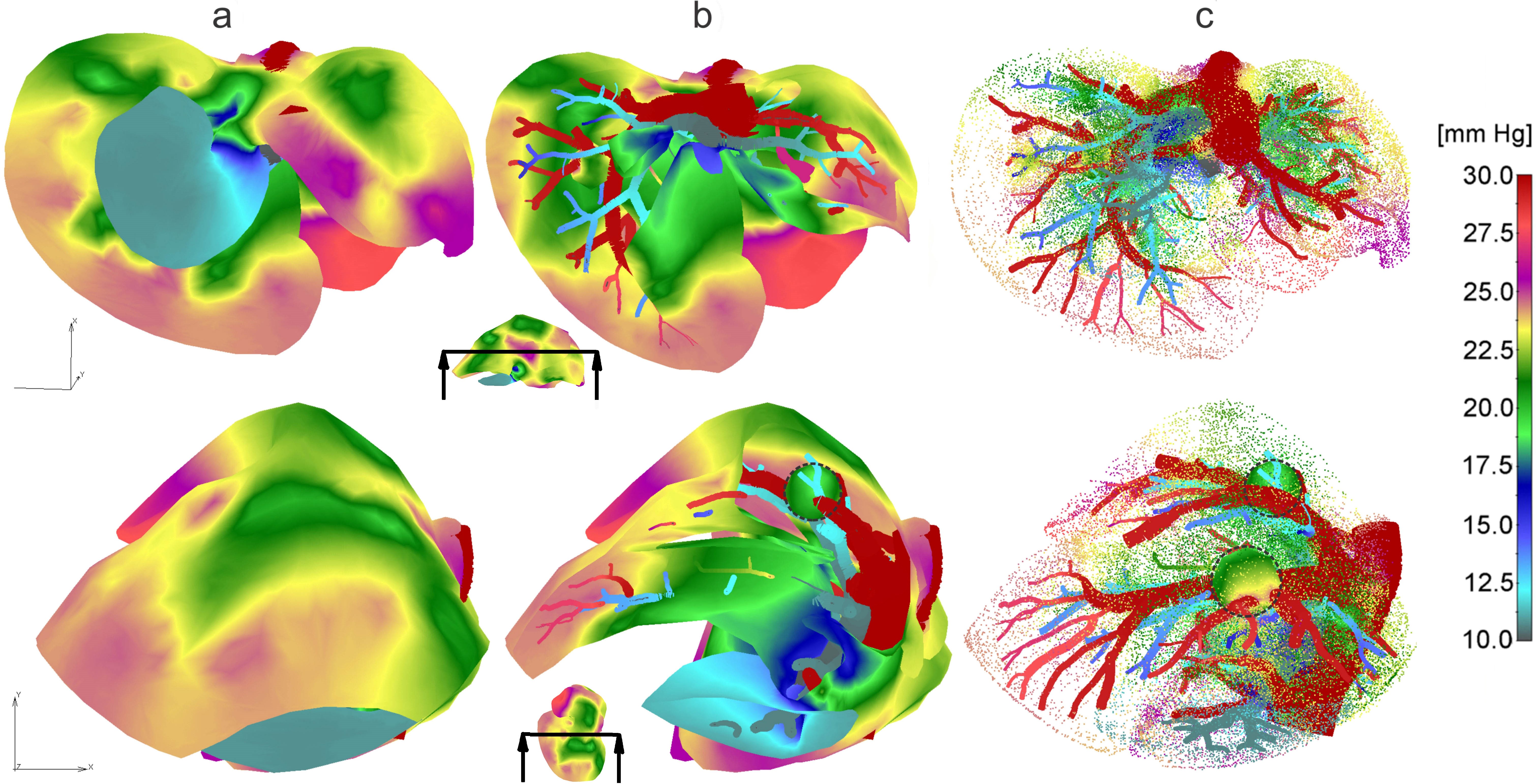
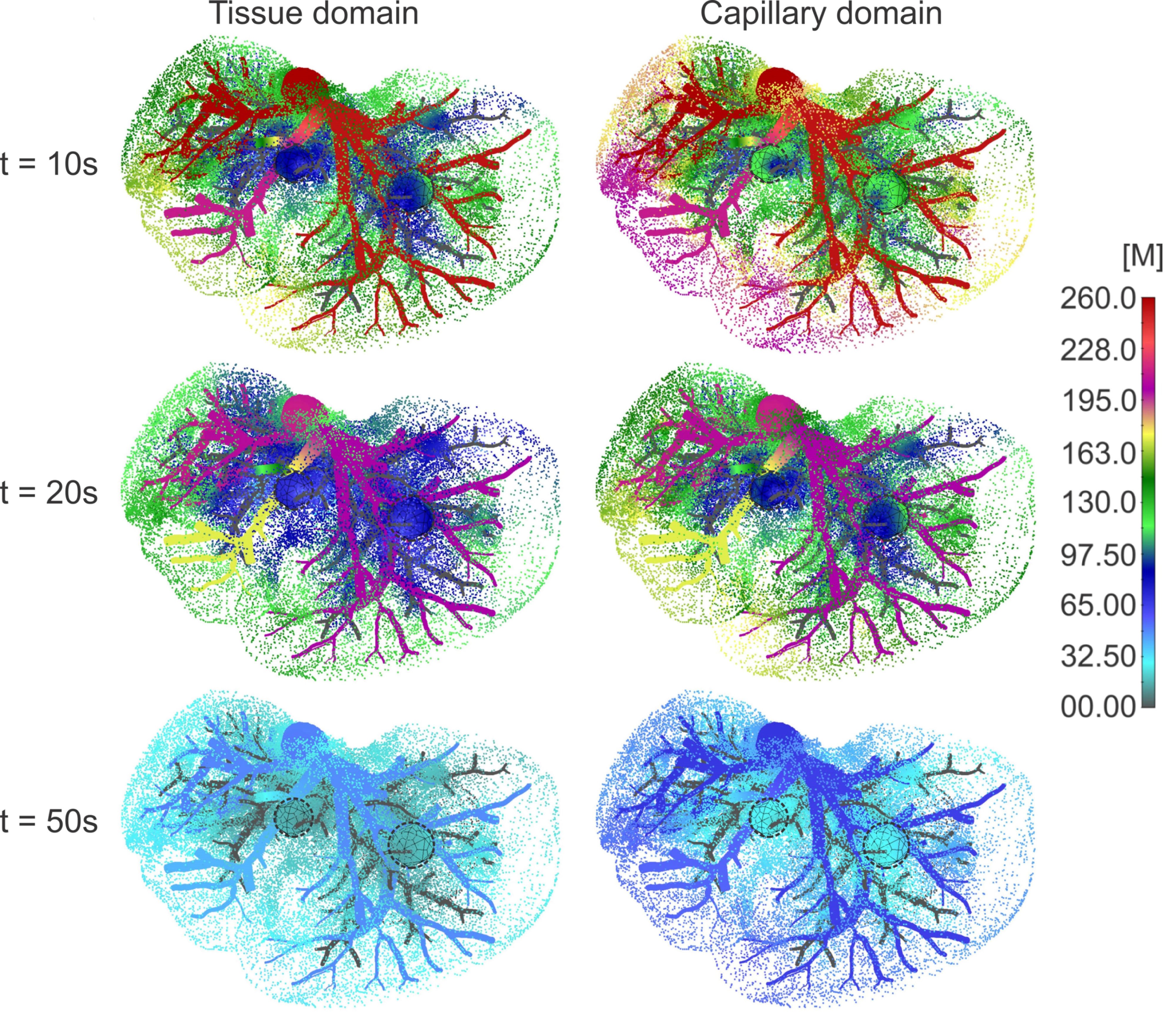
Concentration field in liver with tumors (marked with dashed lines), dotted results in tissue domain and with full mesh in tumors, for times t =10, 20 and 50s.
Pressure fields for two views: a) Full mesh; b) Clipped mesh; c) Dotted representation of results in tissue, and with full mesh in tumors.
The model geometry, including large vessels and tissue, was generated at the Bioengineering R&D Center BIORC in Serbia, according to imaging data from: E. J. Koay, MD Anderson Cancer Center, Houston.
Model of pancreas, with large vessels and finite elements on the surface. Color in blood vessels corresponds to pressures.
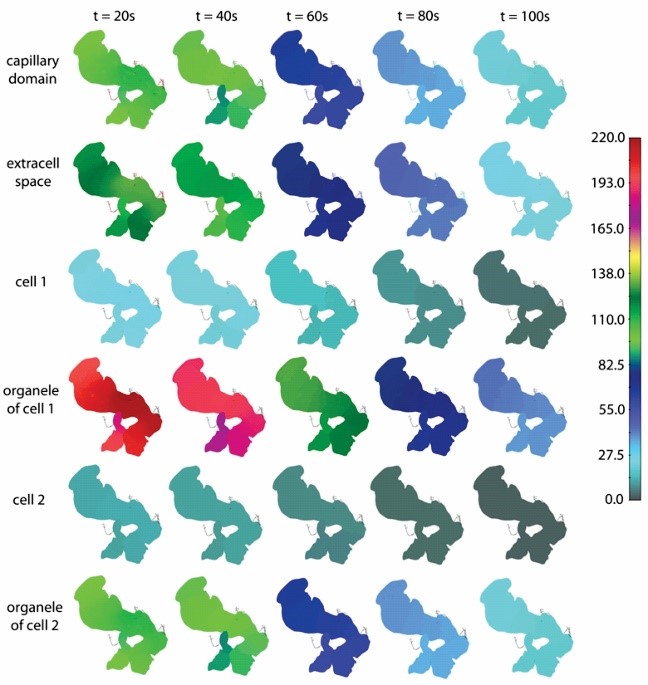
Concentration field within elements CSFE at the pancreas surface, for five selected time points. Maximum concentration, among the shown five time points, at each domain at t=20s when concentration Cin reaches maximum
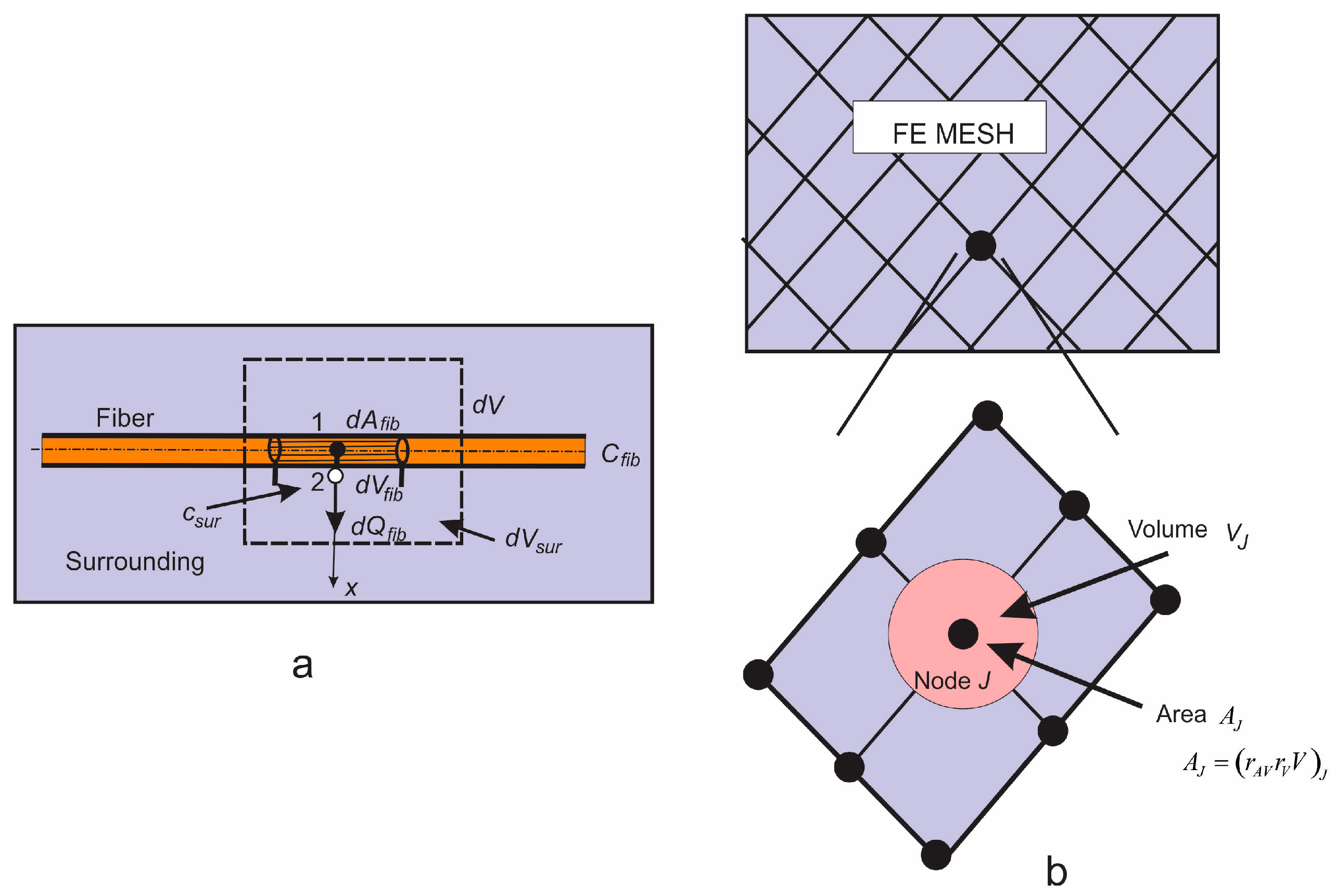
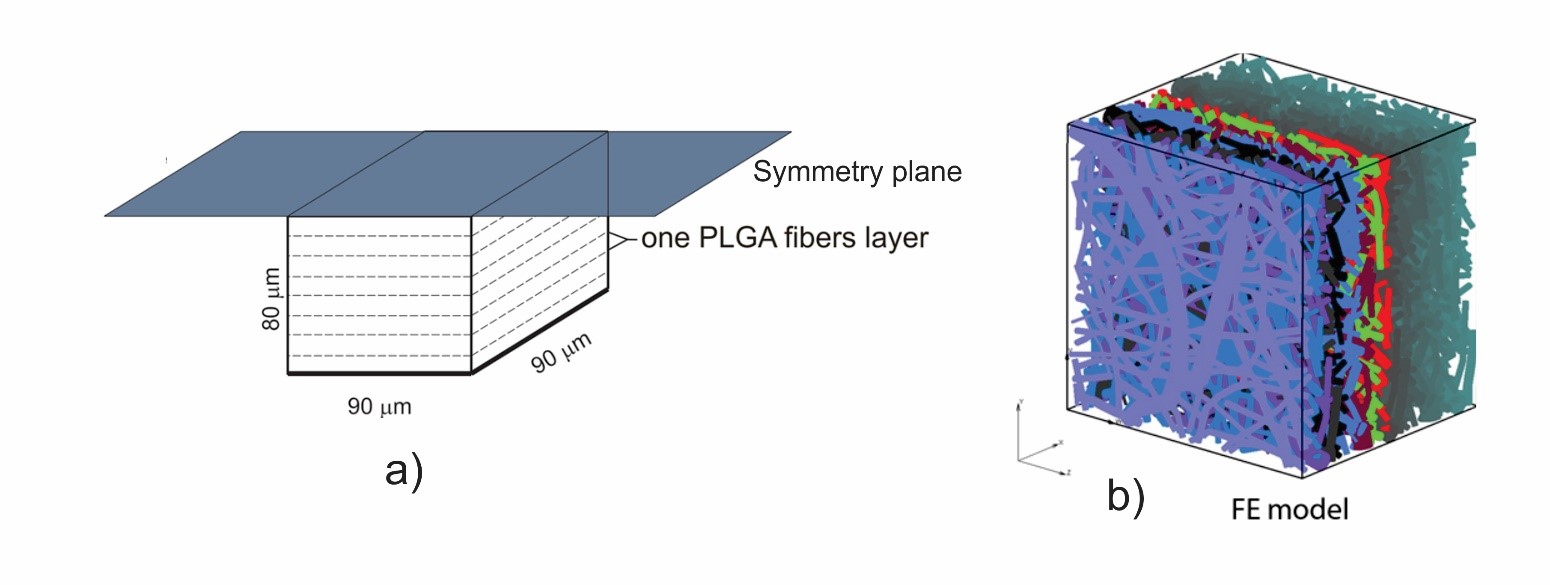
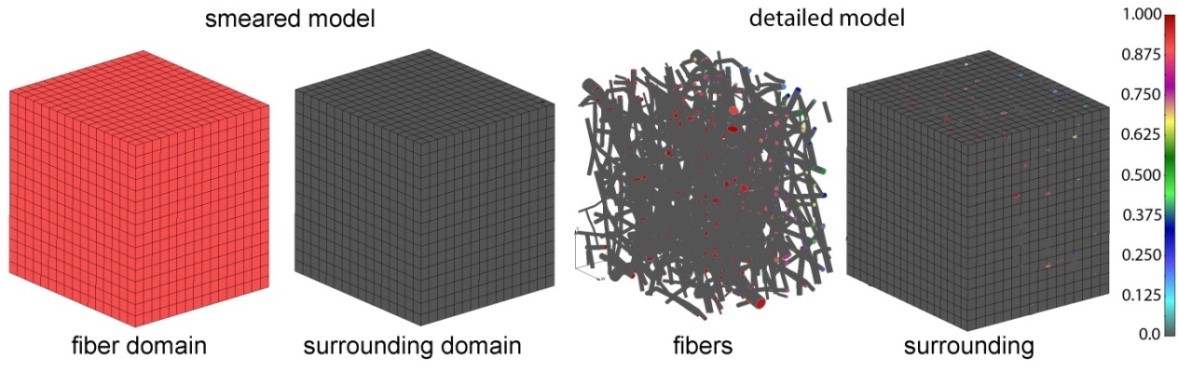
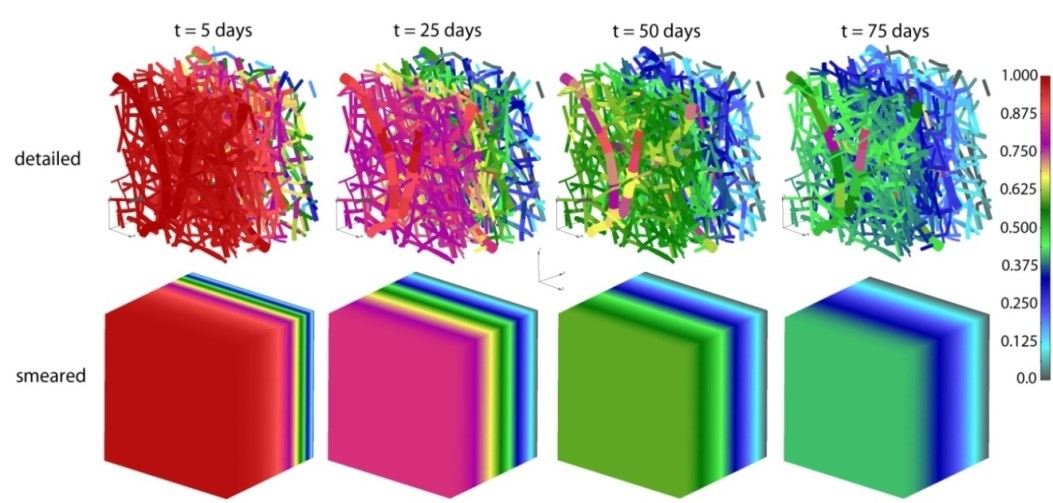
Two different computational models are generated in order to simulate a drug transport from PLGA1 and PLGA2 implants: (a) a detailed FE model with 1D radial elements and (b) a composite smeared finite element model with two different domains: fiber domain and surrounding domain. For the purpose of the detailed FE model, the network of fibers is reconstructed using indoor software from an SEM image of drug loaded PLGA fibers with dimensions 90 µm × 90 µm. By randomly duplicating and displacing the generated layer of 1D fibers into the longitudinal direction of the modeling domain, we can generate a mat of fibers within any implant. Further, assuming symmetric conditions, we can model just one half of the implant. It is also reasonable to adopt a homogenous distribution (or repetition) of one small domain of the fibers, through which we can model just one part of the implant. Thus, the dimensions of our FE models are: 80 µm × 90 µm × 90 µm. The 3D FE mesh (40 × 48 × 48 divisions) consists 64,512 nodes and 36,864 elements; while the number of radial 1D elements is around 7580.
FE model of PLGA implant. a) The 3D domain used in the model, with symmetry plane; b) Generated FE model using a SEM imaging sample.
PLGA domain modeled using a smeared composite finite element or detailed model with the mesh of fibers.
PLGA implant—concentration field in the fibers, for the detailed and smeared model, for the diffusion of Span-80/RhB complex within the PLGA implant.
More details about examples can be found in the Examples directory.