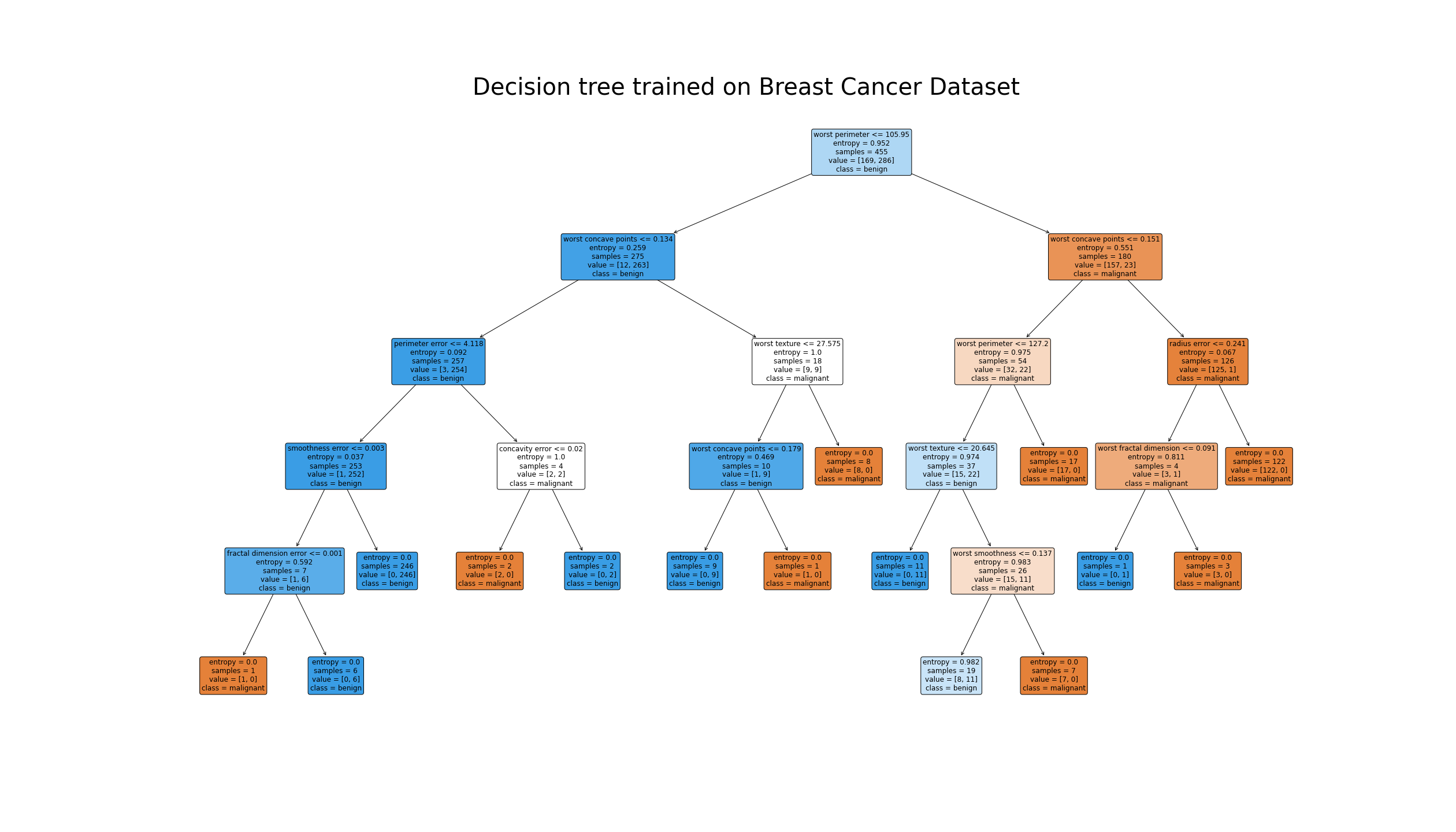
breast cancer dataset from sklearn library.
- malignant: cancer
- benign: not cancer
Breast cancer wisconsin (diagnostic) dataset
--------------------------------------------
Data Set Characteristics:
:Number of Instances: 569
:Number of Attributes: 30 numeric, predictive attributes and the class
:Attribute Information:
- radius (mean of distances from center to points on the perimeter)
- texture (standard deviation of gray-scale values)
- perimeter
- area
- smoothness (local variation in radius lengths)
- compactness (perimeter^2 / area - 1.0)
- concavity (severity of concave portions of the contour)
- concave points (number of concave portions of the contour)
- symmetry
- fractal dimension ("coastline approximation" - 1)
The mean, standard error, and "worst" or largest (mean of the three
worst/largest values) of these features were computed for each image,
resulting in 30 features. For instance, field 0 is Mean Radius, field
10 is Radius SE, field 20 is Worst Radius.
- class:
- WDBC-Malignant
- WDBC-Benign
:Summary Statistics:
===================================== ====== ======
Min Max
===================================== ====== ======
radius (mean): 6.981 28.11
texture (mean): 9.71 39.28
perimeter (mean): 43.79 188.5
area (mean): 143.5 2501.0
smoothness (mean): 0.053 0.163
compactness (mean): 0.019 0.345
concavity (mean): 0.0 0.427
concave points (mean): 0.0 0.201
symmetry (mean): 0.106 0.304
fractal dimension (mean): 0.05 0.097
radius (standard error): 0.112 2.873
texture (standard error): 0.36 4.885
perimeter (standard error): 0.757 21.98
area (standard error): 6.802 542.2
smoothness (standard error): 0.002 0.031
compactness (standard error): 0.002 0.135
concavity (standard error): 0.0 0.396
concave points (standard error): 0.0 0.053
symmetry (standard error): 0.008 0.079
fractal dimension (standard error): 0.001 0.03
radius (worst): 7.93 36.04
texture (worst): 12.02 49.54
perimeter (worst): 50.41 251.2
area (worst): 185.2 4254.0
smoothness (worst): 0.071 0.223
compactness (worst): 0.027 1.058
concavity (worst): 0.0 1.252
concave points (worst): 0.0 0.291
symmetry (worst): 0.156 0.664
fractal dimension (worst): 0.055 0.208
===================================== ====== ======
:Missing Attribute Values: None
:Class Distribution: 212 - Malignant, 357 - Benign
:Creator: Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian
:Donor: Nick Street
:Date: November, 1995
This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets. https://goo.gl/U2Uwz2
Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass.
They describe characteristics of the cell nuclei present in the image.
import pickle
import numpy as np
import pandas as pd
# load trained model to use with your data
def load_model():
return pickle.load(open("./assets/model.pkl", "rb"))
# load model
TREE = load_model()
# load dataframe
DF = pd.read_csv("./assets/cancer.csv")
# get feature names
feature_names = DF.columns[:-1]
features = np.ones((1, len(feature_names)))
# user input
for i, name in enumerate(feature_names):
min_val = DF[name].min()
max_val = DF[name].max()
# print range for each features
print(f"{i + 1:02}.) {name} = [{min_val:.2f}, {max_val:.2f}]")
data = input(f"Enter your {name}: ").strip()
features[0, i] = data if data else DF[name].mean()
# input : 2D array (1, 30)
features = features.astype(np.float64)
prediction = TREE.predict(features)
# output : 2D array
if prediction[0] == 0:
print("\nMalignant, You got a cancer.")
else:
print("\nBenign, You have no cancer.")