code repository for ICA-SAMv7
Internal carotid artery (ICA) stenosis is a life-threatening occult disease. Utilizing CT to examine vascular lesions such as calcified and non-calcified plaques in cases of carotid artery stenosis is a necessary clinical step in formulating the correct treatment plan.
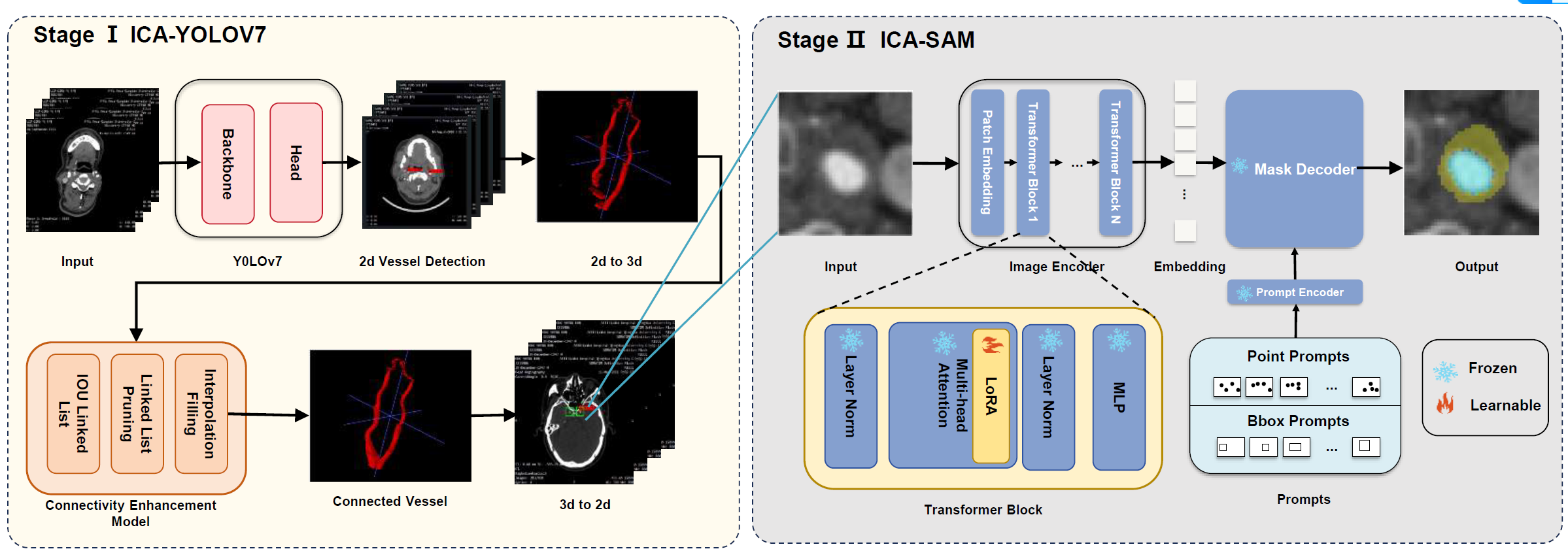
Segment Anything Model (SAM) has shown promising performance in image segmentation tasks, but it performs poorly for carotid artery segmentation. Due to the small size of the calcification and the overlapping between the lumen and calcification. These challenges lead to issues such as mislabeling and boundary fragmentation, as well as high training costs. To address these problems, we propose a two-stage Internal Carotid Artery lesion segmentation method called ICA-SAMv7, which performs coarse and fine segmentation based on the YOLOv7 and SAM model.
Specifically, in the first stage (ICA-YOLOv7), we utilize YOLOv7 for coarse vessel recognition, introducing connectivity information to improve accuracy and achieve precise localization of small target artery vessels. In the second stage (ICA-SAM), we enhance SAM through data augmentation and an efficient parameter fine-tuning strategy. This improves the segmentation accuracy of fine-grained substances in blood vessels while saving training costs. Ultimately, the accuracy of lesion segmentation under the SAM model was increased from the original 48.62% to 82.55%. Extensive comparative experiments have demonstrated the outstanding performance of our algorithm.
Whole Structure of Our Method. In stage Ⅰ, we use YOLOv7 to make coarse artery detection and use the Connectivity Enhancement Model to add connectivity information to improve the accuracy of YOLO segmentation result. In stage Ⅱ, we use LoRA to finetune SAM so that we can get a suitable model for our project.
Clone the repository locally.
git clone https://github.com/BessiePei/ICA-SAMv7.gitCreate the conda env. The code requires python>=3.7, as well as pytorch>=1.7 and torchvision>=0.8. Please follow the instructions here to install both PyTorch and Torchvision dependencies. Installing both PyTorch and TorchVision with CUDA is strongly recommend.
conda create -n ICA-SAMv7 python=3.9
conda activate ICA-SAMv7cd ./ICA-YOLOv7Download pretrained weights from yolov7_weights.pth.
Prepare dataset for the project as VOC dataset structure. Use python voc_annotation.py to get dataset list.
python train.pyChange the model_path in yolo.py to the result weights file path.
python predict.pypython connectivity_enhance.pycd ./ICA-SAMDownload pretrained weights from ica-sam.pth.
python train.pypython test.pyThis project is released under the Apache 2.0 license.
- We thank all medical workers for preparing dataset for our work.
- Thanks to the open-source of the following projects: YOLOv7, Segment Anything , SAM-Med2D.