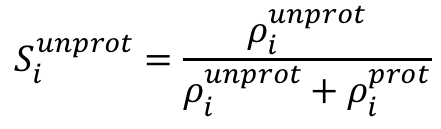The purpose of the CpHMD-Analysis tool is to aid users in processing lambda and log files from Constant pH Molecular Dynamics (CpHMD) simulations run from the molecular dynamics software packages in Amber and CHARMM. This python library is equipped to calculate the unprotonated fraction, pure and mixed states, running unprotonated fractions, and pKas of titratable residues from the lambda files produced by CpHMD simulations. Additionally, functions have been added to the library to make plotting the running unprotonated fractions and titrations curves quick and easy. Since CpHMD is often used with a pH-based replica-exchange protocol the library also can calculate exchange statics and plot the replica walks.
This is the python library and is composed of 4 classes and 2 functions.
1.) class lambda_data
This class allows you to load lambda files and calculate the unprotonated fractions. Additionally, you can compute the running unprotonated fractions over the simulation time.
2.) class plotting_lambda_data
This class allows for the plotting of the running unprotonated fractions over time and titration curves of titratable residues.
3.) class log_analysis_charmm
This class allows for the loading, processing, and plotting of CHARMM (version >C40) formatted log files, resulting in replica-exchange statistics and plotting the replica walk through pH space.
4.) class log_analysis_amber
Does the same thing as the "class log_analysis_charmm," but for Amber formatted log files.
This jupyter notebook is a step-by-step walk through of how to load, process, and plot your lambda and log files. This juptyer notebook can be used as a starting point for your CpHMD project.
The included jupyter notebook can be run on the amber_example_data, which is provided in two directories sample_1 and sample_2. In the directory "sample_1" there is a set of lambda files and a log file from an initial run of pH-based replica exchange of CpHMD using implicit solvent (solvent model: GBNeck2) and "sample_2" has a set of lambda files and a log file from the restart of the simulation. This simulation was conducted on a small test case protein BBL (PDBid: 1W4H).
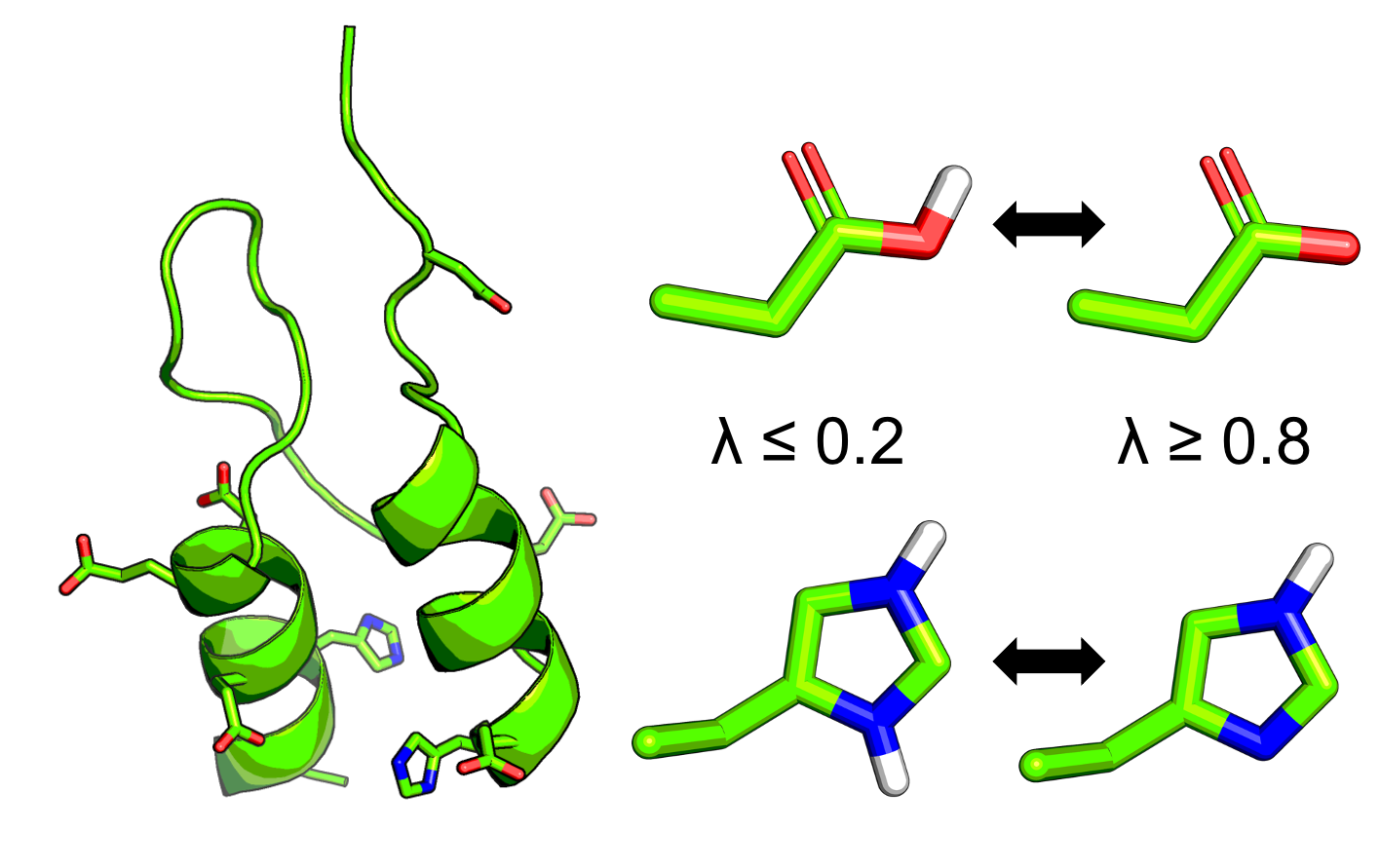
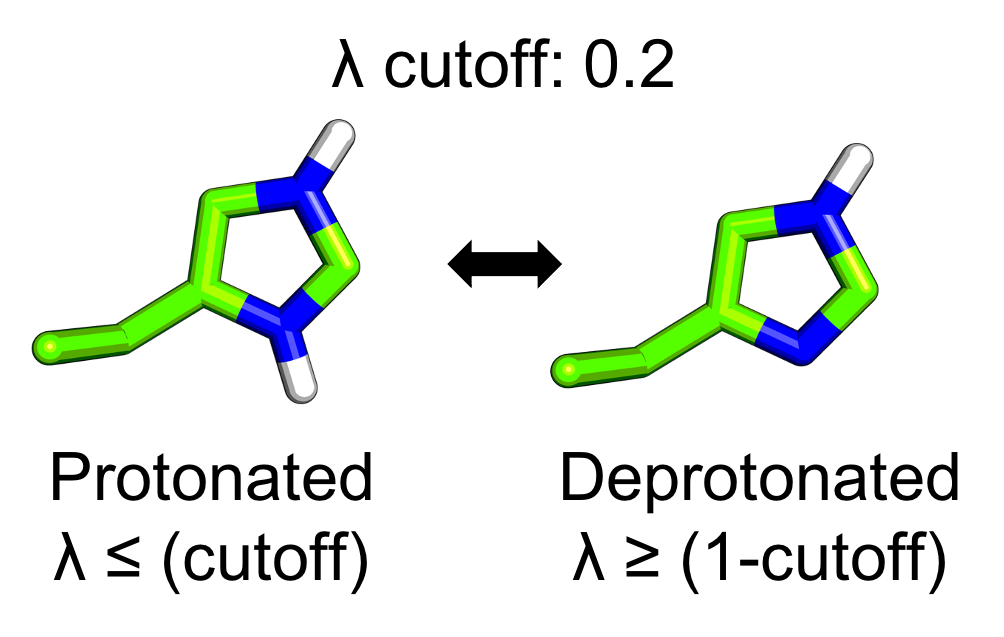
In CpHMD simulations each titratable site, i, is treated as a continuous titration coordinate λi, which is bound between 0 and 1, to govern the progress of that titratable site's protonation/deprotonation. In practice, the λ-value does not always sample exactly either 0 or 1, so a cutoff of 0.2 is used to define whether a given titratable site is protonated or deprotonated. The cutoff value described here is the default cutoff value for all calculations in the CpHMD-Analysis python library.
Using the λ-values with the stated cutoff from a simulation at a single pH we can calculate the unprotonated fraction for each titratable site, Siunprot.
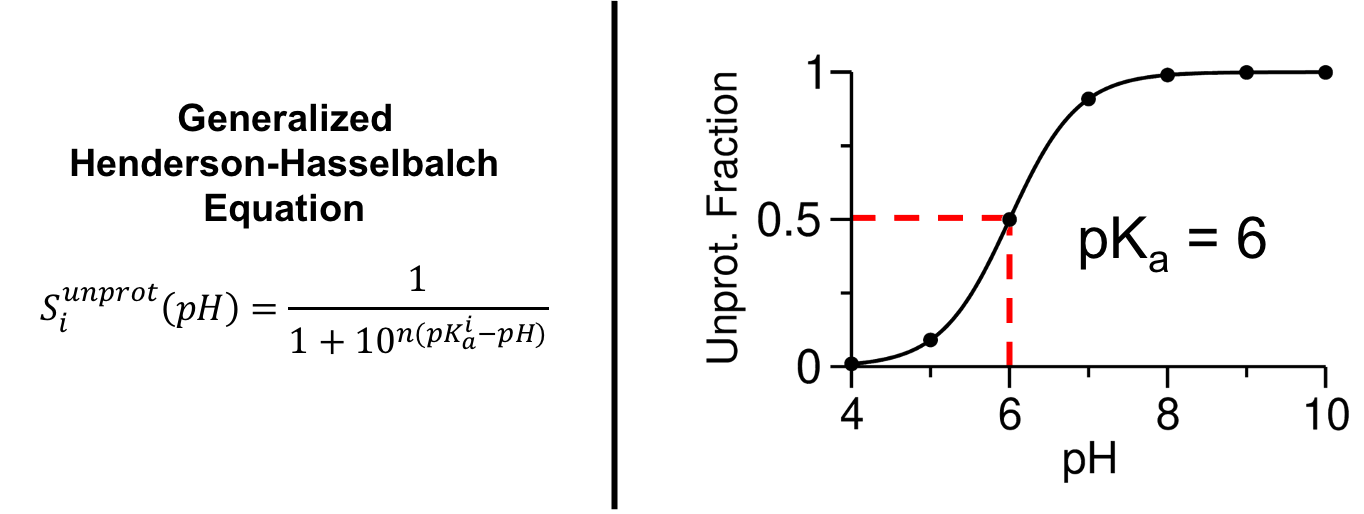
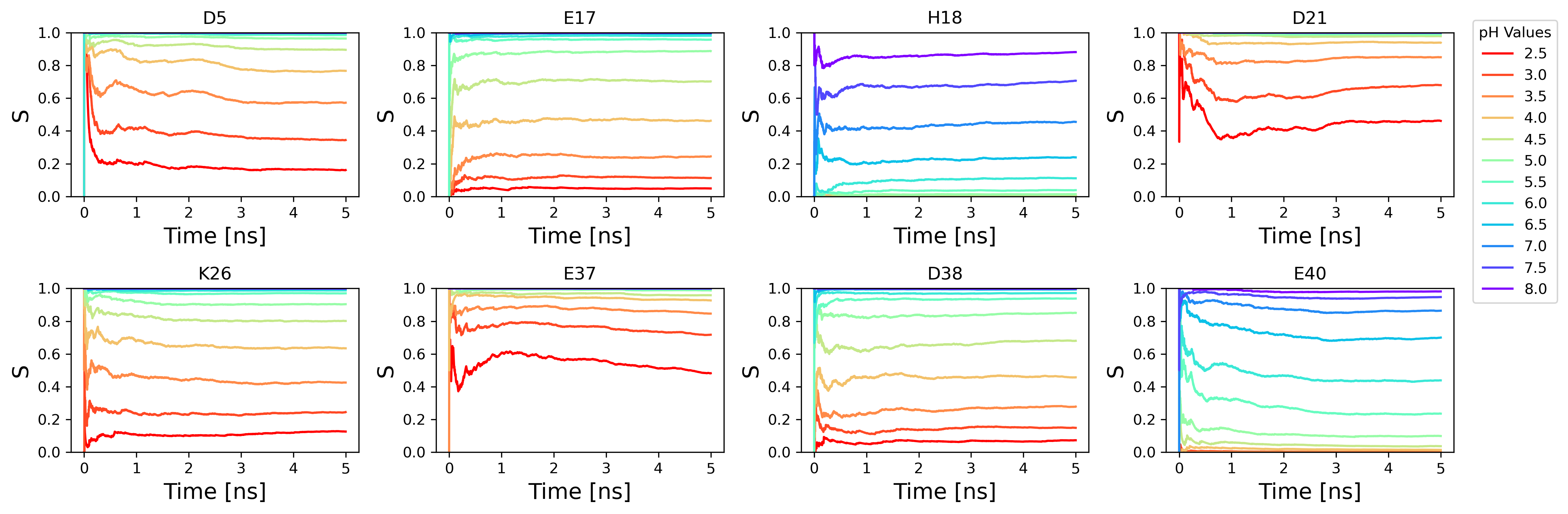
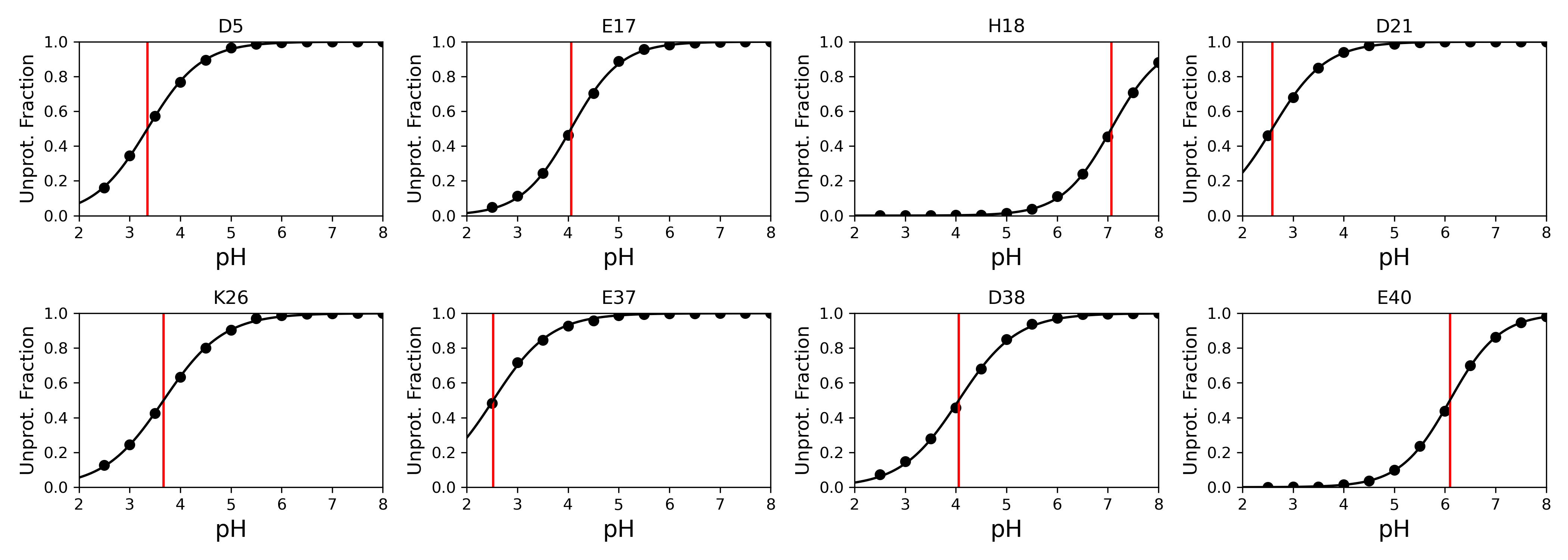
Where ρiunprot and ρiprot are the probabilities that a given titratable site is either unprotonated or protonated, respectively. Keep in mind that this calculation excludes λ-values in a mixed state, meaning the λ-value falls between 0.2 and 0.8. Calculating the Siunprot as a function of time provides a good indication of the convergence of the titratable sites' protonation state in a CpHMD simulation, an example of this is shown in the jupyter notebook tutorial (CpHMD_Analysis_Example_AMBER.ipynb). By calculating the Siunprot at each simulated pH and fitting these points to the generalized Henderson-Hasselbalch equation the pKa can be calculated.
In the Henderson-Hasselbalch equation n is the Hill coefficient, which represents the slope of the transition region of the titration curve. One detail that you might be wondering about is the handling of the tautomeric state of the titratable residues such as Histidine, Aspartic Acid, and Glutamic Acid. In these cases an additional coordinate, χ, is used. Like the λ-value, the χ-value also continuously moves between 0 and 1 representing one or the other tautomeric states and is defined with a cutoff of 0.2. For greater detail on λ- and χ-values please read the articles in the further reading section.
For a step-by-step walk through on how to reproduce the following examples please see the 'CpHMD_Analysis_Example_AMBER.ipynb' jupyter notebook.
This is an example showing the running unprotonated fraction at each pH over time. This type of plot is commonly used to check the convergence of the unprotonated fractions and quality of the pKa values. After the lambda files have been loaded and processed with CpHMD-Analysis library these plots can be generated simply with the following command.
plotting.plot_running_s(ph_objects, phs, resids, titles, xlabel='Time [ns]', steps_to_time_conversion=(1/1000))
Here is an example of the titration curve plots. The red line indicates the pKa. Again, plots like this can be easily made with the following command.
plotting.plot_titration_curves(ph_objects, phs, resids, titles, xrange=[2,8])
Using the following command, the pKas can be output after the lambda files have been processed.
pkas = pka(phs, ph_objects)
| Resids | pKas | Hill Coefficients |
|---|---|---|
| 5 | 3.4 | 0.82 |
| 17 | 4.1 | 0.89 |
| 18 | 7.1 | 0.89 |
| 21 | 2.6 | 0.82 |
| 37 | 3.7 | 0.74 |
| 38 | 2.5 | 0.77 |
| 40 | 4.1 | 0.76 |
| 42 | 6.1 | 0.89 |
Constant-pH Molecular Dynamics Using Continuous Titration Coordinates
Constant-pH Molecular Dynamics with Proton Tautomerism
Continuous Constant pH Molecular Dynamics in Explicit Solvent with pH-based Replica Exchange
For a full range of articles on the development and usage of CpHMD please visit the Shen Lab website.