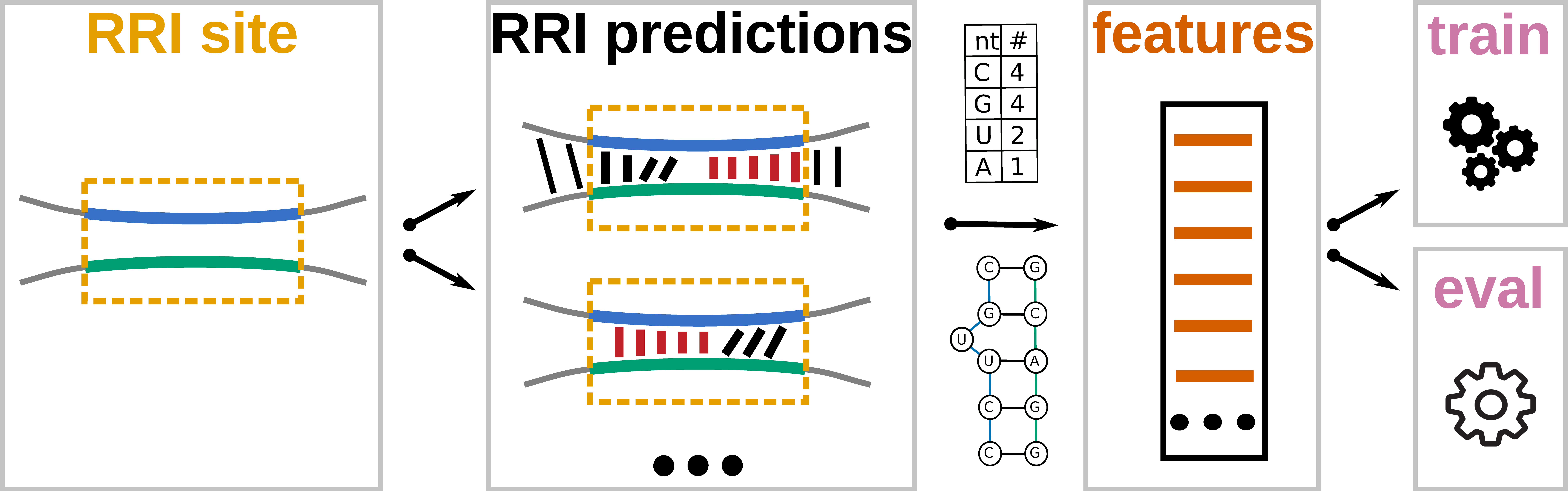
CheRRI detects functional RNA-RNA interaction (RRI) sites, by evaluating if an interaction site most likely occurs in nature. It helps to filter interaction sites generated either experimentally or by an RRI prediction algorithm by removing false positive interactions.
It is an open source project hosted on GitHub.
For help please have a look at our documentation
CheRRI can be run in two modes, the model generation or train mode, or the RRI evaluation or eval mode. For the evaluation of a given set of RRI sites, a model must be specified in CheRRI's eval mode. Here pre-trained models can be applied or the user trains a model using CheRRI's train mode. To train a novel model, an RNA-RNA interactome dataset specifying all RRI sites should be provided. CheRRI makes use of replicate data by checking if an RRI site can be found in all replicates within an overlap threshold. This is how CheRRI builds the set of trusted RRIs. In eval mode, the interaction positions are reformatted in the same way as the trusted RRIs. In both modes, CheRRI uses the same core method to generate a feature set which will be used to select a model in train mode, and in eval mode for the evaluation of the biological relevance of the submitted RRI sites.
CheRRI can be easly istalled via conda. Please have a look at the documentation
If you have any questions you can create an Issue or sent us an e-mail