Important
If you are a BRaVa analyst looking to run these steps in your biobank/cohort, check out these helpful templates:
You'll need to replace filepaths, column names etc in the commands with the corresponding column names in your data. Free text portions of the commands to be changed are placed in square brackets [like this], portions of the commands where you'll need to make a choice between a collection of options are placed in braces {like this}.
Tip
Here's a walkthrough of a single trait and chromosome 11 for all three steps
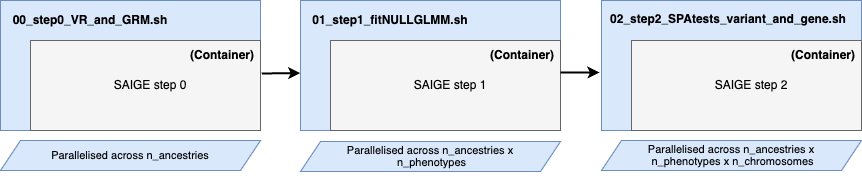
Run SAIGE preprocessing and steps 1 and 2 without any hassle.
- Containerised SAIGE (Docker / Singularity) ✅
- Supporting VCF and PLINK exome data formats ✅
- Parallelised across ancestry, phenotypes and chromosomes ✅
- Sanity checks ✅
- Internet connection (only needed once for download_resources.sh)
- Docker OR Singularity
- Linux OR Mac
To run universal-saige we need to download plink and the SAIGE image. These steps are separated out into download_resources.sh:
bash download_resources.sh --saige-image --plink
bash download_resources.sh --saige-image --plink --singularity
You should now have all the relevant software installed to run all three steps.
- WES data in PLINK (
.bim/.bed/.fam) or VCF format (.gz compressed) - Sample IDs, (ancestry specific)
- SAIGE annotation file (details found here)
- BRaVa phenotype file (tsv) with 'IID' (sample ID) column and covariates
- Genotyping array data for every sample included in the WES data above. Recommended.
Take genotyping array data in plink format, or {WES, WGS} files in {vcf, plink} format, and generate variance ratios and a sparse GRM.
usage: 00_step0_VR_and_GRM.sh
required:
--geneticDataDirectory: directory containing the genetic data (genotyping array data inplinkformat, or{WES, WGS}files in{vcf, plink}format).--geneticDataFormat: format of the genetic data{vcf, plink}. VCF files must be gzipped with.vcf.gzfile extensions.--sampleIDs:.famfile of the sample IDs that are present in the{WES, WGS}data. Note, if this is not all of the samples in the{WES, WGS}dataset, the{WES, WGS}data must be filtered to these samples before running step 1
optional:
-o,--outputPrefix: output prefix from this program (SAIGE step 0) to be used as SAIGE step 1 input.-s,--isSingularity(default:false): is singularity available? If not, it is assumed that docker is available.--generate_GRM(default: false): generate GRM for the genetic data.--generate_plink_for_vr(default: false): generate plink file for vr.
Important
All files contained within --geneticDataDirectory of the type flagged by --geneticDataFormat will be globbed, so please ensure that this contains all of the autosomes for just one biobank/cohort and not multiple!
usage: 01_step1_fitNULLGLMM.sh
required:
-t,--traitType: type of the trait{quantitative, binary}.--genotypePlink: variance ratio plink filename prefix of.bim/.bed/.famfiles. This must relative to the current working directory. Note that samples will be restricted to samples present within the plink.famfile.--sparseGRM: filename of the sparseGRM.mtxfile (output from step 0). This must be relative to the current working directory.--sparseGRMID: filename of the sparseGRM ID file (output from step 0). This must be relative to the current working directory.--phenoFile: filename of the phenotype file. This must be relative to the working directory.--phenoCol: the column names of the phenotype to be analysed in the file specified in--phenoFile.
optional:
-o,--outputPrefix: output prefix from this program (SAIGE step 1) to be used as SAIGE step 2 input.-s,--isSingularity: (default: false): is singularity available? If not, it is assumed that docker is available.-c,--covarColList: comma separated column names (e.g.age,pc1,pc2) of continuous covariates to include as fixed effects in the file specified in--phenoFile. Recall, proposed pilot fixed effect covariates areage,age2,sex,age*sex,age2*sex,PCs.--categCovarColList: comma separated column names of categorical variables to include as fixed effects in the file specified in --phenoFile.--sampleIDCol(default: IID): column containing the sample IDs in the phenotype file, which must match the sample IDs in the plink files.
usage: 02_step2_SPAtests_variant_and_gene.sh
required:
--chr: chromosome to test.--testType: type of test{variant,group}.-p,--plink: plink filename prefix of.bim/.bed/.famfor WES (or WGS restricted to exons). These must be relative to the current working directory.--vcfvcf exome file. If a set of plink files for the WES (or WGS restricted to exons) is not available then this vcf file will be used. This must be present in the current working directory.--modelFile: filename of the model file output from SAIGE step 1. This must be relative to the current working directory.--varianceRatio: filename of the varianceRatio file output from SAIGE step 1. This must be relative to the current working directory.--sparseGRM: filename of the sparseGRM.mtxfile output from SAIGE step 0. This must be relative to the current working directory.--sparseGRMID: filename of the sparseGRM ID file output from SAIGE step 0. This must be relative to the current working directory.
optional:
-o,--outputPrefix: output prefix from this program (SAIGE step 2).-s,--isSingularity(default: false): is singularity available? If not, it is assumed that docker is available.-g,--groupFile: required if group test is selected. Filename of the annotation file used for group tests. This must be in relation to the working directory.--annotations: required if group test is selected. The collection of annotations in the group file to be tested. Please usepLoF,damaging_missense_or_protein_altering,other_missense_or_protein_altering,synonymous,pLoF:damaging_missense_or_protein_altering,pLoF:damaging_missense_or_protein_altering:other_missense_or_protein_altering:synonymous
usage: 03_estimate_nGlmm.sh
required:
--binaryPhenos: space separated list of binary phenotypes.--contPhenos: space separated list of continuous phenotypes.--phenoFile: filename of the phenotype file. This must be relative to, and contained within, the current working directory.--sparseGRM: filename of the sparseGRM .mtx file. This must be relative to, and contained within, the current working directory.--sparseGRMID: filename of the sparseGRM ID file. This must be relative to, and contained within, the current working directory.