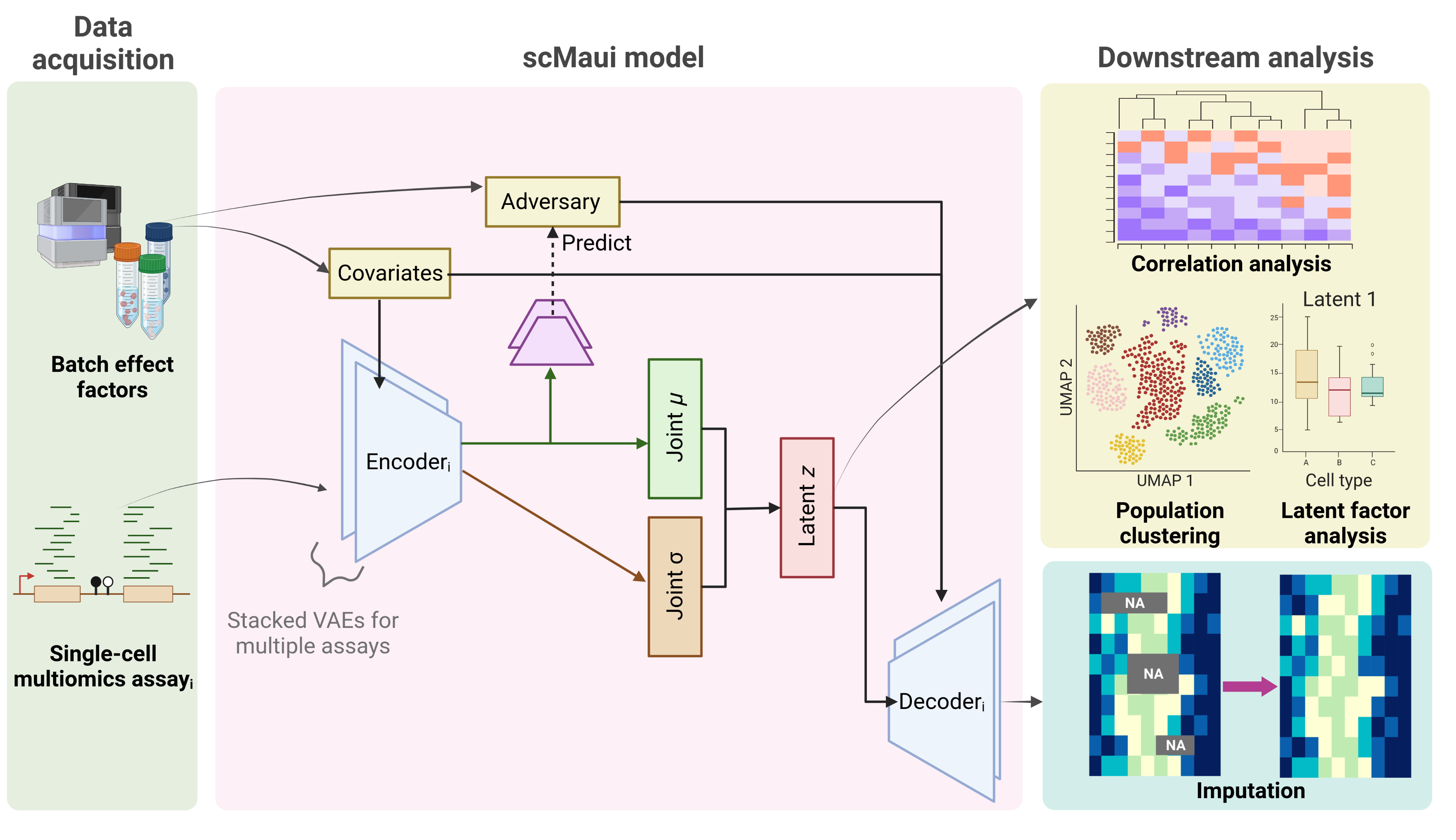
scMaui is python package that implements a variational auto-encoder for multi-omics data integration. The model is capable of handling variable numbers input and output modalities as well as missing modalities. The model also features a range of log-likelihood implementations for determining the reconstruction loss, including the negative binomial or the negative multinomial model.
The package is freely available under a GNU Lesser General Public License v3 or later (LGPLv3+)
scMaui paper is now available on bioRxiv.
pip install scmauiimport pkg_resources
from scmaui.data import load_data, SCDataset
from scmaui.utils import get_model_params
from scmaui.ensembles import EnsembleVAE
# get some toy data
toy_data_path = pkg_resources.resource_filename('scmaui', 'resources/gtx.h5ad')
adatas = load_data([toy_data_path], names=['gtx'])
dataset = SCDataset(adatas, losses=['negbinom'])
# create an scMaui model
params = get_model_params(dataset)
ensemble = EnsembleVAE(params=params)
# fit the model
ensemble.fit(dataset, epochs=10)
ensemble.summary()
# obtain latent features
latents, _ = ensemble.encode(dataset)
# obtain an imputation
imputed = ensemble.impute(dataset)
# obtain input feature attributions
selected_cells = latents.index.tolist()[:5] # select first 5 cells
explanation = ensemble.explain(dataset, cellids=selected_cells)scMaui offers a command line interface for model fitting. The results are stored in an output directory (-output).
scmaui -data adata.h5ad \
-datanames gtx \
-output <outputdir> \
-epochs 200 \
-ensemble_size 10 \
-nlatent 15 \
-adversarial label1 label2 \
-conditional covariate1 covariate2 \
-losses negmul negbinomAdditional information on available hyper-parameters are available through
scmaui -hWe provide tutorials for data preprocessing, scMaui usage and some downstream analyses. You can find the tutorials here.