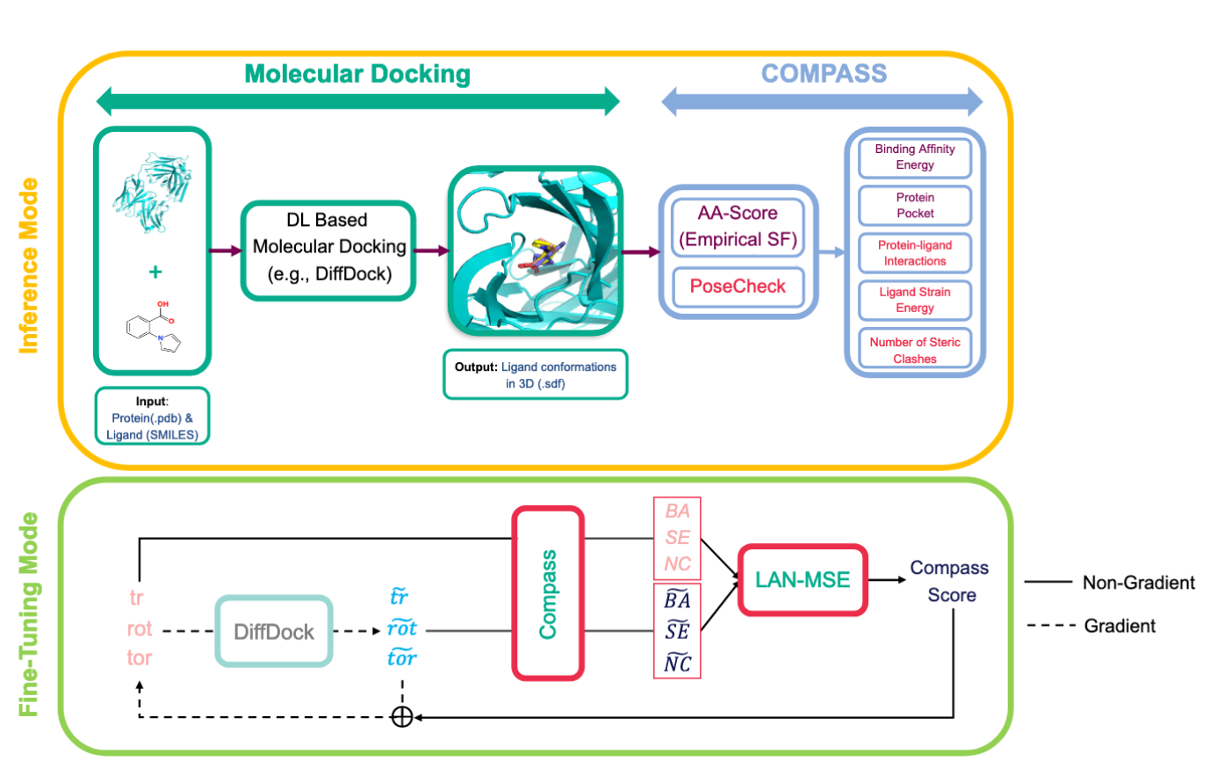
Compass 🧭: A Comprehensive Tool for Accurate and Efficient Molecular Docking in Inference and Fine-Tuning
Navigating Future Drugs with Compass 🧭
Official Implementation of Compass: A Comprehensive Tool for Accurate and Efficient Molecular Docking in Inference and Fine-Tuning paper.
Developed by Ahmet Sarıgün*, Vedran Franke, and Altuna Akalin, Compass is designed for accurate and efficient molecular docking in both inference and fine-tuning phases. This repository provides the necessary code and instructions to utilize the method effectively.
Should you have any questions or encounter issues, please feel free to open an issue on this repository or contact us directly at Ahmet.Sariguen@mdc-berlin.de.
Check out our paper below for more details:
Compass: A Comprehensive Tool for Accurate and Efficient Molecular Docking in Inference and Fine-Tuning,
Ahmet Sarıgün, Vedran Franke, Altuna Akalin
Arxiv, 2024
Set up your development environment using Anaconda. Start by cloning the repository:
git clone https://github.com/BIMSBbioinfo/Compass.gitOnce you have cloned the repository, navigate to its root directory and execute the following commands to create and activate the compass environment:
conda env create --file environment.yml
conda activate compassFor additional details on managing conda environments, refer to the conda documentation.
Our approach for inference aligns with the method used in DiffDock. The same data formats are applicable here as well.
For protein inputs, you can use .pdb files or provide sequences that will be folded using ESMFold. For the ligands, inputs can be in the form of a SMILES string or files readable by RDKit, such as .sdf or .mol2.
To process a single complex, specify the protein using --protein_path protein.pdb or --protein_sequence GIQSYCTPPYSVLQDPPQPVV, and the ligand using --ligand_description ligand.sdf or --ligand_description "COc(cc1)ccc1C#N".
If you want to do a redocking with recursion, you can use --max_recursion_step.
And you are ready to run inference for compass with single complex:
python -W ignore -m main_inference --config DiffDock/default_inference_args.yaml --protein_path example/proteins/1a46_protein_processed.pdb --ligand_description "C1=CN=C(N1)CCNC(=O)CCCC(=O)NCCC2=NC=CN2" --out_dir results/user_predictions_small --max_recursion_step 2You will get Binding Affinity Energy, Strain Energy of Ligand, Number of Steric Clashes of Complex and Interaction Information of Complex. Also, you'll get the protein pocket in .pdb in pockets/ where you save your results in --out_dir to better understand the region of docked molecule in protein pocket.
If you have multiple protein target files and multiple ligand files/SMILES you want to run, give protein files' direction with --protein_dir and indicate the range of them with --protein_start and --protein_end. Also if you have .txt file containing SMILES, you can give the direction with --smiles_dir and range them with --smiles_start and --smiles_end.
Now you can run a couple of proteins and ligands at the same inference run:
python -W ignore -m main_inference --config DiffDock/default_inference_args.yaml --protein_dir example/proteins --smiles_dir example/smiles.txt --out_dir results/user_predictions_small --max_recursion_step 1 --protein_start 0 --protein_end 2 --smiles_start 0 --smiles_end 2Only the PDBBind dataset is utilized in this project. The data processing guidelines provided in DiffDock and the steps for generating ESM Embeddings are also applicable here.
After generating ESM embeddings, run the Inference Mode once to download the pretrained DiffDock-L. Now, we're ready to finetune DiffDock with Compass:
python -W ignore -m finetune --config experiments/model_parameters.ymlplease cite the following paper if you use this code/repository in your research:
@article{sarigun2024compass,
title={Compass: A Comprehensive Tool for Accurate and Efficient Molecular Docking in Inference and Fine-Tuning},
author={Sarigun, Ahmet and Franke, Vedran and Akalin, Altuna},
journal={arXiv preprint arXiv:2406.06841},
year={2024}
}
This code is available for non-commercial scientific research purposes as will be defined in the LICENSE file which is Attribution-NonCommercial-NoDerivatives 4.0 International. By downloading and using this code you agree to the terms in the LICENSE. Third-party datasets and software are subject to their respective licenses.
Components of the code of the spyrmsd by Rocco Meli (MIT license), DiffDock by Gabriele Corso (MIT license), AA-Score by Xiaolin Pan (GNU General Public License v2.0) and PoseCheck by Charlie Harris (MIT license) were integrated in the repo.
We extend our deepest gratitude to the following teams for open-sourcing their valuable Repos:
- DiffDock Team (version 2023 & 2024),
- AA-score Team,
- PoseCheck Team