Deep-Learning-Based Segmentation of Small Extracellular Vesicles in Transmission Electron Microscopy Images
Author of the notebook: Estibaliz Gómez de Mariscal
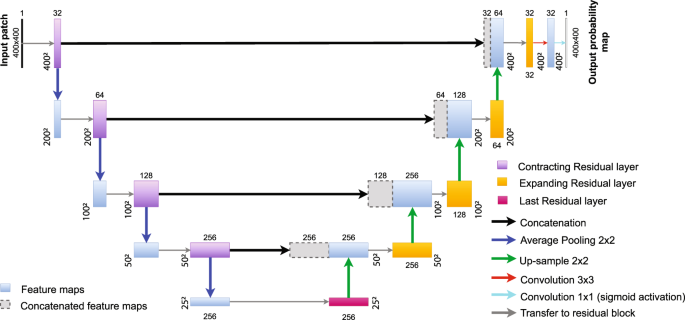
CNN architecture: Fully residual U-Net (FRUNet)
References
If you use this notebook, please cite the original paper:
- Estibaliz Gómez-de-Mariscal, Martin Maška, Anna Kotrbová, Vendula Pospíchalová, Pavel Matula, Arrate Muñoz-Barrutia. Deep-Learning-Based Segmentation of Small Extracellular Vesicles in Transmission Electron Microscopy Images. Scientific Reports, 9(1):13211, 2019.
You can access the original paper here
Data
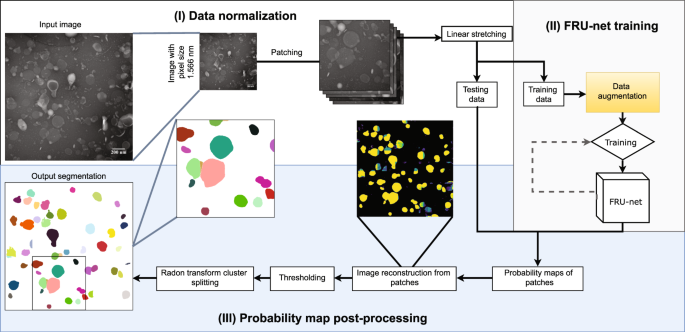
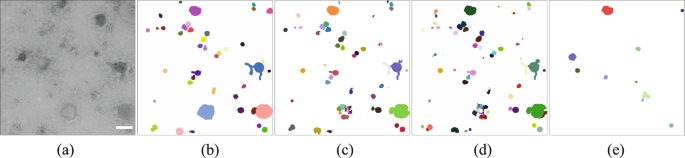
The example data consists of transmission electron microscopy (TEM) images of small extracellular vessicles (sEV). Further information can be found at CBIA Massaryk University.
Use the code
We provide a Jupyter Notebook that can be directly used in Google Colab:
The notebook allows you to:
- Reproduce the results in the paper.
- Fine-tune one of the trained models. Note that you should work on addapting your data if you wish to do so.
Issues
For any question or problem running the code, please use GitHub issues