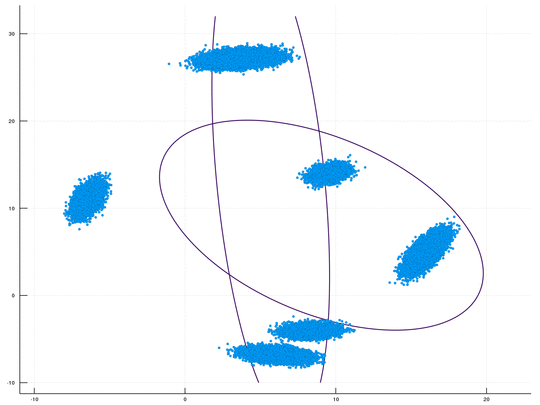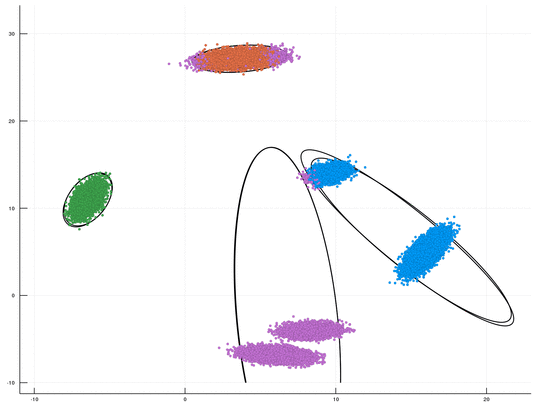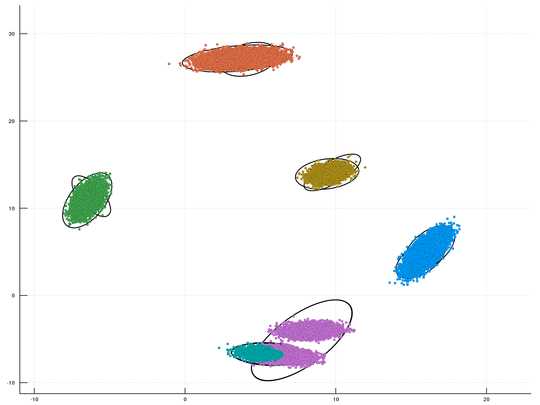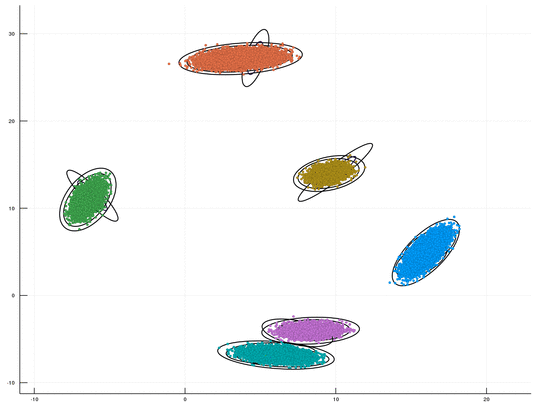
This is a fast GPU implenentation, using CUDA/C++, for sampling-based inference in a Dirichlet Process Mixture Model (DPMM). An optional Python wrapper is also available. This package is Cross Platform (supporting both Windows & Linux). The underlying algorithm is the DPMM sampler proposed by Chang and Fisher III, NIPS 2013. This repository, together with its CPU counterpart, is part of our upcoming paper "CPU- and GPU-based Distributed Sampling in Dirichlet Process Mixtures for Large-scale Analysis" (authors: Dinari*, Zamir*, Fisher III, and Freifeld).
This package was developed and tested with C++14 and CUDA 11.2 on Windows 10 (Visual Studio 2019), Ubuntu 18.04, and Ubuntu 21.04. The following dependencies are required:
- GPU
- CUDA driver 11.2 or higher
- OpenCV - for visualization purposes (i.e., plotting points in 2D)
- Install CUDA version 11.2 (or higher) from https://developer.nvidia.com/CUDA-downloads
- git clone https://github.com/BGU-CS-VIL/DPMMSubClusters_GPU
- Add Environment Variables:
- Add "CUDA_VERSION" with the value of the version of your CUDA installation (e.g., 11.5).
- Make sure that CUDA_PATH exist. If it is missing add it with a path to CUDA (e.g., export CUDA_PATH=/usr/local/cuda-11.5/).
- Make sure that the relevant CUDA paths are included in $PATH and $LD_LIBRARY_PATH (e.g., export PATH=/usr/local/cuda-11.5/bin:$PATH, export LD_LIBRARY_PATH=/usr/local/cuda- 11.5/lib64:$LD_LIBRARY_PATH).
- Add "CUDA_VERSION" with the value of the version of your CUDA installation (e.g., 11.5).
- Make sure that CUDA_PATH exists. If it is missing add it with a path to CUDA (e.g., C:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.5)
- Install cmake if necessary.
- For Windows only (optional, used on for debugging purposes): Install OpenCV
- run Git Bash
- cd <YOUR_PATH_TO_DPMMSubClusters_GPU>/DPMMSubClusters
- ./installOCV.sh
For Windows for the CUDA/C++ package both of the build options below are viable. For Linux use Option 2.
- Option 1: DPMMSubClusters.sln - Solution file for Visual Studio 2019
- Option 2: CMakeLists.txt
- Run in the terminal:
cd <PATH_FOR_DPMMSubClusters_DIRECTORY> mkdir build cd build cmake -S ../ - Build:
- Windows: cmake --build . --config Release --target ALL_BUILD
- Linux: cmake --build . --config Release --target all
- Run in the terminal:
Windows
Linux
Both binaries were compiled with CUDA 11.2, note that you still need to have cuda and cudnn installed in order to use these.
The package currently contains priors for handling Multinomial or Gaussian mixture models.
While being very versatile in the setting and configuration, there are 2 modes which you can work with, either the Basic, which will use mostly predefined configuration, and will take the data as an argument, or Advanced use, which allows more configuration, loading data from file.
In order to run in the basic mode while generating sample data, use the code:
srand(12345);
data_generators data_generators;
MatrixXd x;
std::shared_ptr<LabelsType> labels = std::make_shared<LabelsType>();
double** tmean;
double** tcov;
int N = (int)pow(10, 5);
int D = 2;
int numClusters = 2;
int numIters = 100;
data_generators.generate_gaussian_data(N, D, numClusters, 100.0, x, labels,
tmean, tcov);
std::shared_ptr<hyperparams> hyper_params = std::make_shared<niw_hyperparams>(
1.0, VectorXd::Zero(D), 5,
MatrixXd::Identity(D, D));
dp_parallel_sampling_class dps(N, x, 0, prior_type::Gaussian);
ModelInfo dp = dps.dp_parallel(hyper_params, N, numIters, 1, true, false,
false, 15, labels);
- all_data - The data, should be
DxN. - local_hyper_params - The prior you plan to use, can be either Multinomial, or
NIW. - alpha_param - Concentration parameter
- iters - Number of iterations
- verbose - Printing status on every iteration.
- burnout - How many iteration before allowing clusters to split/merge, reducing this number will result in faster inference, but with higher variance between the different runs.
- gt - Ground Truth, if supplied will perform
NMIandVItests on every iteration.
dp_parallel will return the following ModelInfo with few important structures inside it:
labels, weights, iter_count
Note that weights does not sum up to 1, but to 1 minus the weight of the non-instanisated components.
Reducing the burnout will increase the speed and reduce stability, increasing the variance in the results.
When supplied with gt kwarg, it will perform NMI and Variation of Information analysis on each iteration.
There are a few parameters that can be used to run the program. In order to set the hyperparams the params_path parameter can be used. The value of the parameter should be a path for a Json file which includes the hyperparams (i.e. alpha or hyper_params for the prior). In order to use this parameter follow this syntax:
--params_path=<PATH_TO_JSON_FILE_WITH_MODEL_PARAMS>
There are few more parameters like model_path for the path to a npy file which include the model and result_path for the path of the output results.
Our code has support for both Gaussian and Multinomial distributions. It can be easily adapted to other component distributions, e.g., Poisson, as long as they belong to an exponential family. The default distribution is Gaussian. To specify a distribution other than a Gaussian, use the prior_type parameter. For example:
--prior_type="Multinomial"
The Json file containing the model parameters can contain many parameters that can be controlled by the user. A few examples are: alpha, prior, number of iterations, burn_out and kernel. The full list of parameters can be seen in the function init() in global_params. The result file is a Json file which by default contains the predicted labels, the weights, the Normalized Mutual Information (NMI) score and the running time per iteration. A few other parameters can be added to the result file. Samples for these additional parameters are commented out in the main.cpp file.
For any questions: dinari@post.bgu.ac.il or razzam@post.bgu.ac.il
Contributions, feature requests, suggestions etc.. are welcome.
If you use this code for your work, please cite the following work:
@article{dinari2022cpu,
title={CPU-and GPU-based Distributed Sampling in Dirichlet Process Mixtures for Large-scale Analysis},
author={Dinari, Or and Zamir, Raz and Fisher III, John W and Freifeld, Oren},
journal={arXiv preprint arXiv:2204.08988},
year={2022}
}