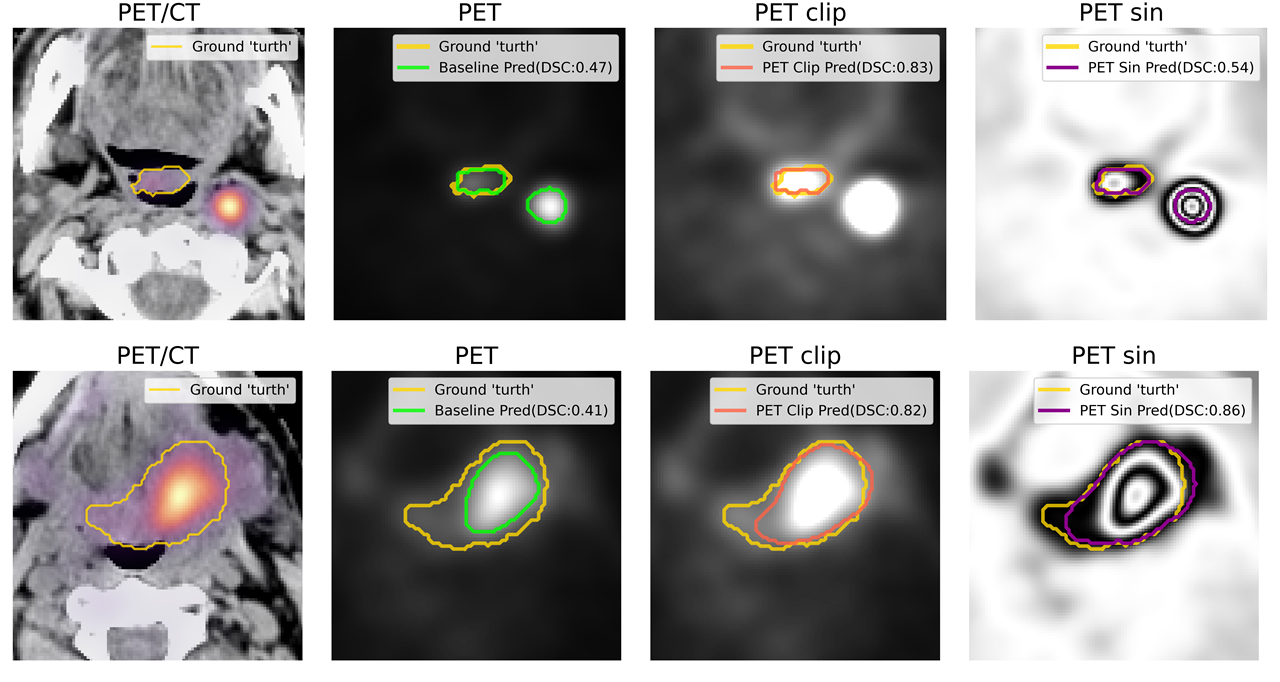
This is a repository for competition of MICCAI 2021: HECKTOR - head and neck gross tumor volume(GTV) segmentation.
Task 1: Segmentation of GTV:
Task 2: Treatment outcomes prediction:
Comparing deep learning and conventional machine learning for outcome prediction of head and neck cancer in PET/CT
HECKTOR public code for resample images to istropical 1mm grid with bounding box(144x144x144):
git clone https://github.com/voreille/hecktor
cd hecktor/src/resampling/
python resample.py
We use nnUNet as the baseline model for the development of GTV multimodality segmentation network. install nnUNet:
git clone https://github.com/MIC-DKFZ/nnUNet.git
cd nnUNet
pip install -e .
convert data:
We've provded a script to convert sampled image to nnUNet format. Please change the your file location for both downloaded_data_dir and downloaded_data_dir_test
if convert train set only:
python data_conversion.py
if conver both train and test set:
python data_conversion.py --test=True
Run evaluation on 5-folds CV using following command for task id XXX:
nnUNet_find_best_configuration -m 3d_fullres -t XXX --strict
Ren, J., Huynh, B. N., Groendahl, A. R., Tomic, O., Futsaether, C. M., & Korreman, S. S. (2021, September). PET Normalizations to Improve Deep Learning Auto-Segmentation of Head and Neck Tumors in 3D PET/CT. In 3D Head and Neck Tumor Segmentation in PET/CT Challenge (pp. 83-91). Springer, Cham.
@inproceedings{ren2021pet,
title={PET Normalizations to Improve Deep Learning Auto-Segmentation of Head and Neck Tumors in 3D PET/CT},
author={Ren, Jintao and Huynh, Bao-Ngoc and Groendahl, Aurora Rosvoll and Tomic, Oliver and Futsaether, Cecilia Marie and Korreman, Stine Sofia},
booktitle={3D Head and Neck Tumor Segmentation in PET/CT Challenge},
pages={83--91},
year={2021},
organization={Springer}
}
and
Huynh, B. N., Ren, J., Groendahl, A. R., Tomic, O., Korreman, S. S., & Futsaether, C. M. (2021, September). Comparing deep learning and conventional machine learning for outcome prediction of head and neck cancer in PET/CT. In 3D Head and Neck Tumor Segmentation in PET/CT Challenge (pp. 318-326). Springer, Cham.
@inproceedings{huynh2021comparing,
title={Comparing deep learning and conventional machine learning for outcome prediction of head and neck cancer in PET/CT},
author={Huynh, Bao-Ngoc and Ren, Jintao and Groendahl, Aurora Rosvoll and Tomic, Oliver and Korreman, Stine Sofia and Futsaether, Cecilia Marie},
booktitle={3D Head and Neck Tumor Segmentation in PET/CT Challenge},
pages={318--326},
year={2021},
organization={Springer}
}