coronavirus 
The coronavirus package provides a tidy format dataset of the 2019 Novel Coronavirus COVID-19 (2019-nCoV) epidemic and the vaccination efforts by country. The raw data is being pulled from the Johns Hopkins University Center for Systems Science and Engineering (JHU CCSE) Coronavirus repository.
More details available
here, and a csv format
of the package dataset available
here
Important Notes
- As this an ongoing situation, frequent changes in the data format may occur, please visit the package changelog (e.g., News) and/or see pinned issues to get updates about those changes
- As of Auguest 4th JHU CCSE stopped track recovery cases, please see this issue for more details
- Negative values and/or anomalies may occurred in the data for the
following reasons:
- The calculation of the daily cases from the raw data which is in cumulative format is done by taking the daily difference. In some cases, some retro updates not tie to the day that they actually occurred such as removing false positive cases
- Anomalies or error in the raw data
- Please see this issue for more details
Vignettes
Additional documentation available on the followng vignettes:
- Introduction to the Coronavirus Dataset
- Covid19R Project Data Format
- Update the coronavirus Dataset
- Covid19 Vaccine Data
- Geospatial Visualization
Installation
Install the CRAN version:
install.packages("coronavirus")Install the Github version (refreshed on a daily bases):
# install.packages("devtools")
devtools::install_github("RamiKrispin/coronavirus")Datasets
The package provides the following two datasets:
-
coronavirus - tidy (long) format of the JHU CCSE datasets. That includes the following columns:
date- The date of the observation, usingDateclassprovince- Name of province/state, for countries where data is provided split across multiple provinces/statescountry- Name of country/regionlat- The latitude codelong- The longitude codetype- An indicator for the type of cases (confirmed, death, recovered)cases- Number of cases on given dateuid- Country codeprovince_state- Province or state if applicableiso2- Officially assigned country code identifiers with two-letteriso3- Officially assigned country code identifiers with three-lettercode3- UN country codefips- Federal Information Processing Standards code that uniquely identifies counties within the USAcombined_key- Country and province (if applicable)population- Country or province populationcontinent_name- Continent namecontinent_code- Continent code
-
covid19_vaccine - a tidy (long) format of the the Johns Hopkins Centers for Civic Impact global vaccination dataset by country. This dataset includes the following columns:
country_region- Country or region namedate- Data collection date in YYYY-MM-DD formatdoses_admin- Cumulative number of doses administered. When a vaccine requires multiple doses, each one is counted independentlypeople_partially_vaccinated- Cumulative number of people who received at least one vaccine dose. When the person receives a prescribed second dose, it is not counted twicepeople_fully_vaccinated- Cumulative number of people who received all prescribed doses necessary to be considered fully vaccinatedreport_date_string- Data report date in YYYY-MM-DD formatuid- Country codeprovince_state- Province or state if applicableiso2- Officially assigned country code identifiers with two-letteriso3- Officially assigned country code identifiers with three-lettercode3- UN country codefips- Federal Information Processing Standards code that uniquely identifies counties within the USAlat- Latitudelong- Longitudecombined_key- Country and province (if applicable)population- Country or province populationcontinent_name- Continent namecontinent_code- Continent code
Data refresh
While the coronavirus CRAN
version is updated
every month or two, the Github (Dev)
version is updated on a
daily bases. The update_dataset function enables to overcome this gap
and keep the installed version with the most recent data available on
the Github version:
library(coronavirus)
update_dataset()Note: must restart the R session to have the updates available
Alternatively, you can pull the data using the
Covid19R project data
standard
format
with the refresh_coronavirus_jhu function:
covid19_df <- refresh_coronavirus_jhu()
head(covid19_df)
#> date location location_type location_code location_code_type data_type value lat long
#> 1 2021-10-04 Afghanistan country AF iso_3166_2 deaths_new 6 33.93911 67.709953
#> 2 2021-10-03 Afghanistan country AF iso_3166_2 deaths_new 0 33.93911 67.709953
#> 3 2020-11-09 Afghanistan country AF iso_3166_2 deaths_new 6 33.93911 67.709953
#> 4 2021-10-10 Afghanistan country AF iso_3166_2 deaths_new 4 33.93911 67.709953
#> 5 2021-10-06 Afghanistan country AF iso_3166_2 deaths_new 6 33.93911 67.709953
#> 6 2020-11-10 Afghanistan country AF iso_3166_2 deaths_new 12 33.93911 67.709953Usage
data("coronavirus")
head(coronavirus)
#> date province country lat long type cases
#> 1 2020-01-22 Alberta Canada 53.9333 -116.5765 confirmed 0
#> 2 2020-01-23 Alberta Canada 53.9333 -116.5765 confirmed 0
#> 3 2020-01-24 Alberta Canada 53.9333 -116.5765 confirmed 0
#> 4 2020-01-25 Alberta Canada 53.9333 -116.5765 confirmed 0
#> 5 2020-01-26 Alberta Canada 53.9333 -116.5765 confirmed 0
#> 6 2020-01-27 Alberta Canada 53.9333 -116.5765 confirmed 0Summary of the total confrimed cases by country (top 20):
library(dplyr)
summary_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases)
summary_df %>% head(20)
#> # A tibble: 20 × 2
#> country total_cases
#> <chr> <int>
#> 1 US 44683014
#> 2 India 34020730
#> 3 Brazil 21597949
#> 4 United Kingdom 8311851
#> 5 Russia 7742899
#> 6 Turkey 7540193
#> 7 France 7164924
#> 8 Iran 5742083
#> 9 Argentina 5268653
#> 10 Spain 4980206
#> 11 Colombia 4975656
#> 12 Italy 4707087
#> 13 Germany 4343591
#> 14 Indonesia 4231046
#> 15 Mexico 3732429
#> 16 Poland 2928065
#> 17 South Africa 2913880
#> 18 Ukraine 2697176
#> 19 Philippines 2690455
#> 20 Malaysia 2361529Summary of new cases during the past 24 hours by country and type (as of 2021-10-13):
library(tidyr)
coronavirus %>%
filter(date == max(date)) %>%
select(country, type, cases) %>%
group_by(country, type) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type,
values_from = total_cases) %>%
arrange(-confirmed)
#> # A tibble: 195 × 4
#> # Groups: country [195]
#> country confirmed death recovered
#> <chr> <int> <int> <int>
#> 1 US 120321 3054 NA
#> 2 United Kingdom 41669 136 NA
#> 3 Turkey 31248 236 NA
#> 4 Russia 27926 962 NA
#> 5 India 18987 246 NA
#> 6 Ukraine 17100 493 NA
#> 7 Romania 15733 390 NA
#> 8 Germany 12317 14 NA
#> 9 Iran 12298 194 NA
#> 10 Thailand 10064 82 NA
#> 11 Malaysia 7950 68 NA
#> 12 Brazil 7852 176 NA
#> 13 Philippines 7083 173 NA
#> 14 Serbia 6699 51 NA
#> 15 Georgia 4837 26 NA
#> 16 Netherlands 3772 13 NA
#> 17 Belgium 3667 13 NA
#> 18 Vietnam 3461 106 NA
#> 19 Bulgaria 3327 98 NA
#> 20 Singapore 3190 9 NA
#> 21 Cameroon 3003 33 NA
#> 22 Italy 2769 37 NA
#> 23 Spain 2758 42 NA
#> 24 Australia 2744 18 NA
#> 25 Lithuania 2740 26 NA
#> 26 Canada 2706 79 NA
#> 27 Poland 2640 40 NA
#> 28 Austria 2614 15 NA
#> 29 Slovakia 2406 20 NA
#> 30 Cuba 2354 28 NA
#> 31 Greece 2312 31 NA
#> 32 Latvia 2236 17 NA
#> 33 Kazakhstan 2084 35 NA
#> 34 Belarus 2060 17 NA
#> 35 Moldova 2052 29 NA
#> 36 Ireland 2051 26 NA
#> 37 Croatia 2022 27 NA
#> 38 Korea, South 1937 13 NA
#> 39 Mongolia 1920 15 NA
#> 40 Iraq 1766 35 NA
#> # … with 155 more rowsPlotting daily confirmed and death cases in Brazil:
library(plotly)
coronavirus %>%
group_by(type, date) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type, values_from = total_cases) %>%
arrange(date) %>%
mutate(active = confirmed - death - recovered) %>%
mutate(active_total = cumsum(active),
recovered_total = cumsum(recovered),
death_total = cumsum(death)) %>%
plot_ly(x = ~ date,
y = ~ active_total,
name = 'Active',
fillcolor = '#1f77b4',
type = 'scatter',
mode = 'none',
stackgroup = 'one') %>%
add_trace(y = ~ death_total,
name = "Death",
fillcolor = '#E41317') %>%
add_trace(y = ~recovered_total,
name = 'Recovered',
fillcolor = 'forestgreen') %>%
layout(title = "Distribution of Covid19 Cases Worldwide",
legend = list(x = 0.1, y = 0.9),
yaxis = list(title = "Number of Cases"),
xaxis = list(title = "Source: Johns Hopkins University Center for Systems Science and Engineering"))Plot the confirmed cases distribution by counrty with treemap plot:
conf_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases) %>%
mutate(parents = "Confirmed") %>%
ungroup()
plot_ly(data = conf_df,
type= "treemap",
values = ~total_cases,
labels= ~ country,
parents= ~parents,
domain = list(column=0),
name = "Confirmed",
textinfo="label+value+percent parent")data(covid19_vaccine)
head(covid19_vaccine)
#> country_region date doses_admin people_partially_vaccinated people_fully_vaccinated report_date_string uid province_state iso2 iso3 code3 fips lat long combined_key population
#> 1 Afghanistan 2021-02-22 0 0 0 2021-02-22 4 <NA> AF AFG 4 <NA> 33.93911 67.709953 Afghanistan 38928341
#> 2 Afghanistan 2021-02-23 0 0 0 2021-02-23 4 <NA> AF AFG 4 <NA> 33.93911 67.709953 Afghanistan 38928341
#> 3 Afghanistan 2021-02-24 0 0 0 2021-02-24 4 <NA> AF AFG 4 <NA> 33.93911 67.709953 Afghanistan 38928341
#> 4 Afghanistan 2021-02-25 0 0 0 2021-02-25 4 <NA> AF AFG 4 <NA> 33.93911 67.709953 Afghanistan 38928341
#> 5 Afghanistan 2021-02-26 0 0 0 2021-02-26 4 <NA> AF AFG 4 <NA> 33.93911 67.709953 Afghanistan 38928341
#> 6 Afghanistan 2021-02-27 0 0 0 2021-02-27 4 <NA> AF AFG 4 <NA> 33.93911 67.709953 Afghanistan 38928341
#> continent_name continent_code
#> 1 Asia AS
#> 2 Asia AS
#> 3 Asia AS
#> 4 Asia AS
#> 5 Asia AS
#> 6 Asia ASPlot the top 20 vaccinated countries:
covid19_vaccine %>%
filter(date == max(date),
!is.na(population)) %>%
mutate(fully_vaccinated_ratio = people_fully_vaccinated / population) %>%
arrange(- fully_vaccinated_ratio) %>%
slice_head(n = 20) %>%
arrange(fully_vaccinated_ratio) %>%
mutate(country = factor(country_region, levels = country_region)) %>%
plot_ly(y = ~ country,
x = ~ round(100 * fully_vaccinated_ratio, 2),
text = ~ paste(round(100 * fully_vaccinated_ratio, 1), "%"),
textposition = 'auto',
orientation = "h",
type = "bar") %>%
layout(title = "Percentage of Fully Vaccineted Population - Top 20 Countries",
yaxis = list(title = ""),
xaxis = list(title = "Source: Johns Hopkins Centers for Civic Impact",
ticksuffix = "%"))
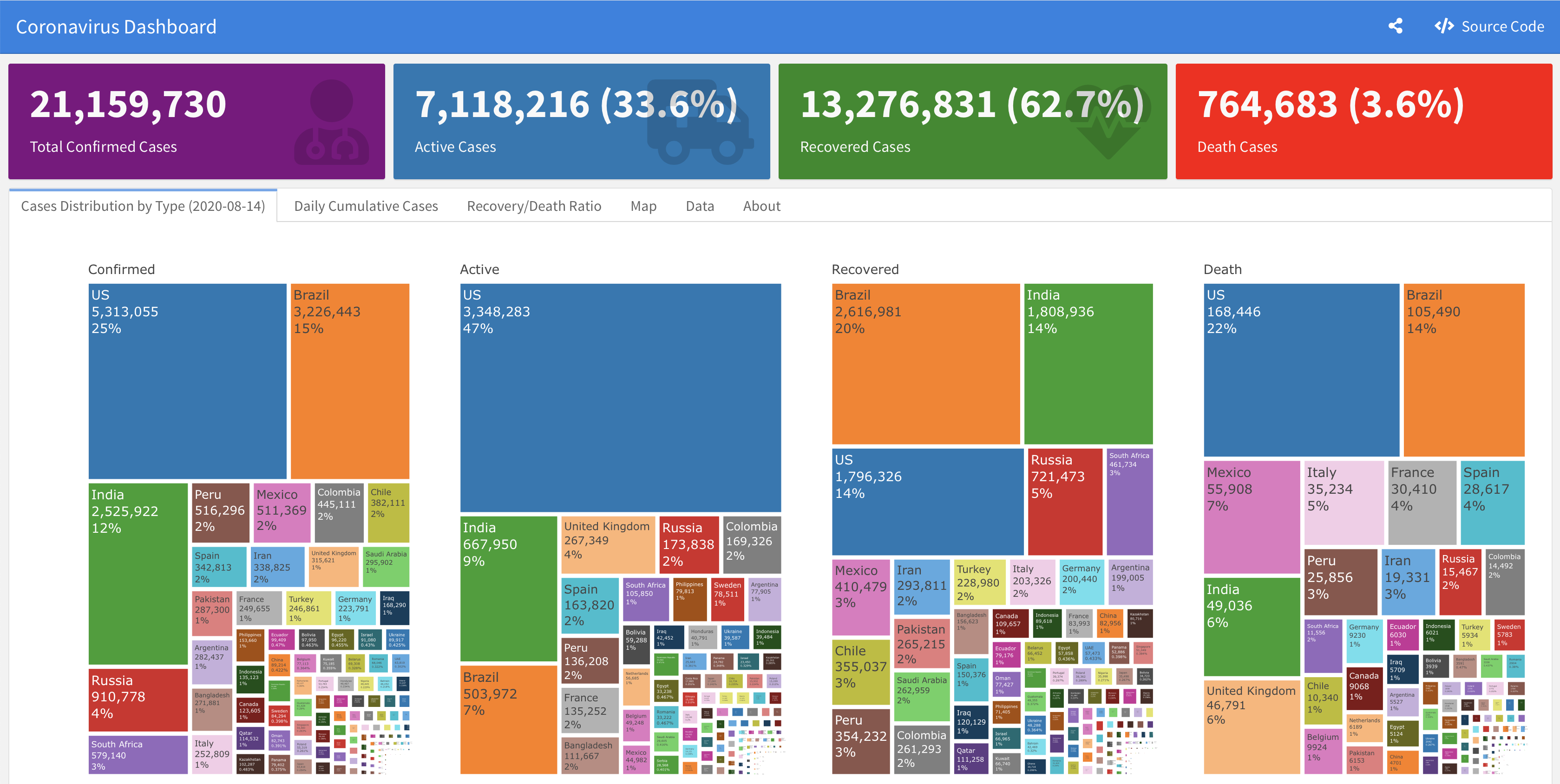
Dashboard
Note: Currently, the dashboard is under maintenance due to recent changes in the data structure. Please see this issue
A supporting dashboard is available here
Data Sources
The raw data pulled and arranged by the Johns Hopkins University Center for Systems Science and Engineering (JHU CCSE) from the following resources:
- World Health Organization (WHO): https://www.who.int/
- DXY.cn. Pneumonia. 2020.
https://ncov.dxy.cn/ncovh5/view/pneumonia.
- BNO News:
https://bnonews.com/index.php/2020/04/the-latest-coronavirus-cases/
- National Health Commission of the People’s Republic of China (NHC):
http:://www.nhc.gov.cn/xcs/yqtb/list_gzbd.shtml - China CDC (CCDC):
http:://weekly.chinacdc.cn/news/TrackingtheEpidemic.htm
- Hong Kong Department of Health:
https://www.chp.gov.hk/en/features/102465.html
- Macau Government: https://www.ssm.gov.mo/portal/
- Taiwan CDC:
https://sites.google.com/cdc.gov.tw/2019ncov/taiwan?authuser=0
- US CDC: https://www.cdc.gov/coronavirus/2019-ncov/index.html
- Government of Canada:
https://www.canada.ca/en/public-health/services/diseases/2019-novel-coronavirus-infection/symptoms.html
- Australia Government Department of
Health:https://www.health.gov.au/news/health-alerts/novel-coronavirus-2019-ncov-health-alert
- European Centre for Disease Prevention and Control (ECDC): https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases
- Ministry of Health Singapore (MOH): https://www.moh.gov.sg/covid-19
- Italy Ministry of Health: https://www.salute.gov.it/nuovocoronavirus
- 1Point3Arces: https://coronavirus.1point3acres.com/en
- WorldoMeters: https://www.worldometers.info/coronavirus/
- COVID Tracking Project: https://covidtracking.com/data. (US Testing and Hospitalization Data. We use the maximum reported value from “Currently” and “Cumulative” Hospitalized for our hospitalization number reported for each state.)
- French Government: https://dashboard.covid19.data.gouv.fr/
- COVID Live (Australia): https://covidlive.com.au/
- Washington State Department of Health:https://www.doh.wa.gov/Emergencies/COVID19
- Maryland Department of Health: https://coronavirus.maryland.gov/
- New York State Department of Health: https://health.data.ny.gov/Health/New-York-State-Statewide-COVID-19-Testing/xdss-u53e/data
- NYC Department of Health and Mental Hygiene: https://www1.nyc.gov/site/doh/covid/covid-19-data.page and https://github.com/nychealth/coronavirus-data
- Florida Department of Health Dashboard: https://services1.arcgis.com/CY1LXxl9zlJeBuRZ/arcgis/rest/services/Florida_COVID19_Cases/FeatureServer/0 and https://fdoh.maps.arcgis.com/apps/opsdashboard/index.html#/8d0de33f260d444c852a615dc7837c86
- Palestine (West Bank and Gaza): https://corona.ps/details
- Israel: https://govextra.gov.il/ministry-of-health/corona/corona-virus/
- Colorado: https://covid19.colorado.gov/data)