Code for the paper "Different Algorithms (Might) Uncover Different Patterns" first presented in the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2023) [ArXiv].
Machine learning techniques are ubiquitous in bioinformatics and EEG research in particular. However, there is still a lot of ambiguity and room for experimental design choices in the experiment setup and analysis pipeline. This raises an important question: Do different experimental design choices, particularly regarding model selection, influence the patterns uncovered in EEG data? Such variations could potentially result in differing, or even contradictory, interpretations.
data: we have provided sample data we used in the paper for demonstration purposes.example_eeg: contains eeg from one subject including eyes opened and closed states.example_subj_EC_raw.fif.gz: sample eeg recording for eyes closed resting state.example_subj_EO_raw.fif.gz: sample eeg recording for eyes opened resting state.example_subj_EC_preproc.pickle: preprocessed eeg data from example_subj_EC_raw.fif.gz.example_subj_EO_preproc.pickle: preprocessed eeg data from example_subj_EO_raw.fif.gz.
example_training_set: contains the12-alltraining set we used in the paper.
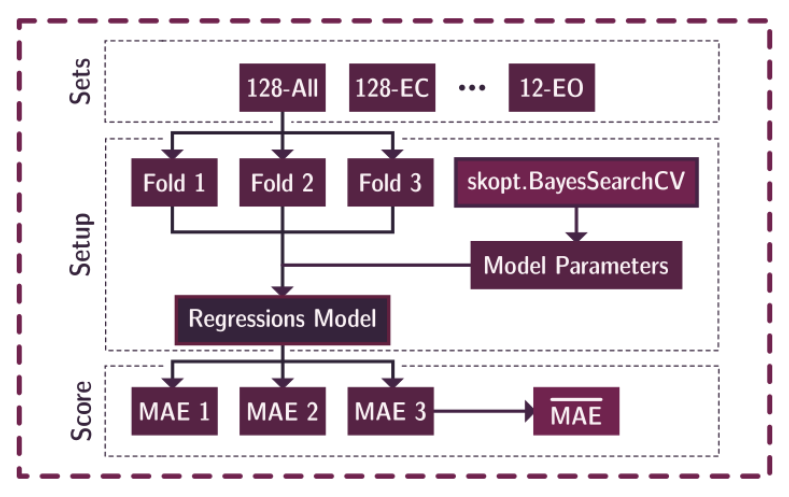
models: contains the corresponding code for each of the 10 models explored in the original paper.training.ipynb: contains the code for training and tuning the models.shap_values.ipynb: contains the code for computing the shap values.shap_values.pickle: is a dict object containing the shap values and fold indexes they were computed on for the specific model.
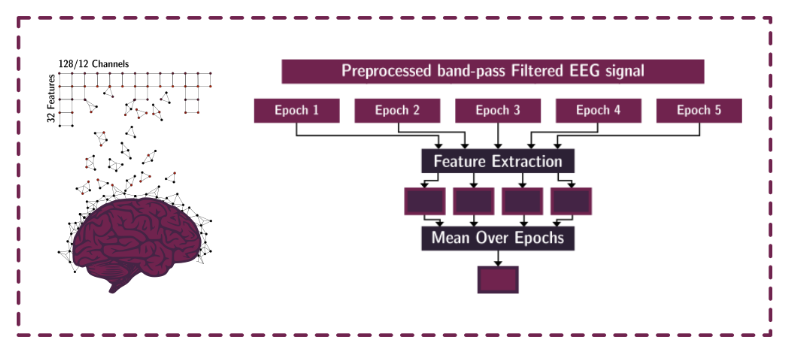
example_eeg_preprocessing.ipynb: a step by step notebook showcasing how to to run our data prerpocessing pipeline.example_eeg_feature_extraction.ipynb: a step by step notebook showcasing how to run our feature extraction pipeline.example_plot_interpolation_map.ipynb: a step by step notebook showcasing how to create the regional interpolation plots.example_aggregated_shap_value.ipynb: a step by step notebook showcasing how to aggregate shap values and create rank orders of the feature groups based on the shap value.example_shap_agreement_metric.ipynb: a step by step notebook showcasing how to compute the ShapAgreement metric between models.
@INPROCEEDINGS{10385662,
author={Ettling, Tobias and Saba-Sadiya, Sari and Roig, Gemma},
booktitle={2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM)},
title={Different Algorithms (Might) Uncover Different Patterns: A Brain-Age Prediction Case Study},
year={2023},
pages={4051-4058},
keywords={Machine learning algorithms;Focusing;Prediction algorithms;Brain modeling;Feature extraction;Robustness;Electroencephalography},
doi={10.1109/BIBM58861.2023.10385662}
}